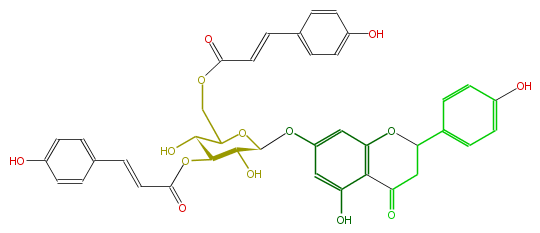
Mol:FL2FAAGS0013
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.9737 -0.6523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9737 -0.6523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9737 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9737 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5300 -1.6159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5300 -1.6159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0863 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0863 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0863 -0.6523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0863 -0.6523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5300 -0.3311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5300 -0.3311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6426 -1.6159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6426 -1.6159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1989 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1989 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1989 -0.6523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1989 -0.6523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6426 -0.3311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6426 -0.3311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7550 -0.3313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7550 -0.3313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3220 -0.6586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3220 -0.6586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8890 -0.3313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8890 -0.3313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8890 0.3234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8890 0.3234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3220 0.6508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3220 0.6508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7550 0.3234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7550 0.3234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6426 -2.1597 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6426 -2.1597 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4174 -0.3311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4174 -0.3311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4560 0.6508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4560 0.6508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5300 -2.2580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5300 -2.2580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6466 -0.4645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6466 -0.4645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2784 -0.9505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2784 -0.9505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7483 -0.7443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7483 -0.7443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2367 -0.7388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2367 -0.7388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6085 -0.3670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6085 -0.3670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0723 -0.6118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0723 -0.6118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1562 -0.7588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1562 -0.7588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8523 -0.9784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8523 -0.9784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4445 -1.2543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4445 -1.2543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4982 0.1260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4982 0.1260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1861 -1.5565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1861 -1.5565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9251 -2.0086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9251 -2.0086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8530 -1.5565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8530 -1.5565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1864 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1864 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8533 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8533 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1206 -0.5160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1206 -0.5160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6552 -0.5160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6552 -0.5160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9225 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9225 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6552 -1.4419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6552 -1.4419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1206 -1.4419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1206 -1.4419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4560 -0.9790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4560 -0.9790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4982 0.7917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4982 0.7917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0874 1.2025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0874 1.2025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3606 1.6758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3606 1.6758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4078 1.2025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4078 1.2025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0682 1.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0682 1.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6111 1.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6111 1.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8808 1.3236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8808 1.3236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4203 1.3236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4203 1.3236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6901 1.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6901 1.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4203 2.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4203 2.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8808 2.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8808 2.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2291 1.7908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2291 1.7908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 31 32 2 0 0 0 0 | + | 31 32 2 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 35 1 0 0 0 0 | + | 40 35 1 0 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 30 42 1 0 0 0 0 | + | 30 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL2FAAGS0013 | + | ID FL2FAAGS0013 |
| − | KNApSAcK_ID C00008213 | + | KNApSAcK_ID C00008213 |
| − | NAME Prunin 3'',6''-di-p-coumarate | + | NAME Prunin 3'',6''-di-p-coumarate |
| − | CAS_RN 84823-66-5 | + | CAS_RN 84823-66-5 |
| − | FORMULA C39H34O14 | + | FORMULA C39H34O14 |
| − | EXACTMASS 726.194855796 | + | EXACTMASS 726.194855796 |
| − | AVERAGEMASS 726.67886 | + | AVERAGEMASS 726.67886 |
| − | SMILES O(C(C2O)C(C(COC(C=Cc(c6)ccc(c6)O)=O)OC(Oc(c5)cc(c(c5O)4)OC(CC4=O)c(c3)ccc(c3)O)2)O)C(=O)C=Cc(c1)ccc(O)c1 | + | SMILES O(C(C2O)C(C(COC(C=Cc(c6)ccc(c6)O)=O)OC(Oc(c5)cc(c(c5O)4)OC(CC4=O)c(c3)ccc(c3)O)2)O)C(=O)C=Cc(c1)ccc(O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
0.9737 -0.6523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9737 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5300 -1.6159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0863 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0863 -0.6523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5300 -0.3311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6426 -1.6159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1989 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1989 -0.6523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6426 -0.3311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7550 -0.3313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3220 -0.6586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8890 -0.3313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8890 0.3234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3220 0.6508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7550 0.3234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6426 -2.1597 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4174 -0.3311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4560 0.6508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5300 -2.2580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6466 -0.4645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2784 -0.9505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7483 -0.7443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2367 -0.7388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6085 -0.3670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0723 -0.6118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1562 -0.7588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8523 -0.9784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4445 -1.2543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4982 0.1260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1861 -1.5565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9251 -2.0086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8530 -1.5565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1864 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8533 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1206 -0.5160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6552 -0.5160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9225 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6552 -1.4419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1206 -1.4419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4560 -0.9790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4982 0.7917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0874 1.2025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3606 1.6758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4078 1.2025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0682 1.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6111 1.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8808 1.3236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4203 1.3236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6901 1.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4203 2.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8808 2.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2291 1.7908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
18 1 1 0 0 0 0
14 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 18 1 0 0 0 0
26 30 1 0 0 0 0
28 31 1 0 0 0 0
31 32 2 0 0 0 0
31 33 1 0 0 0 0
33 34 2 0 0 0 0
34 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 35 1 0 0 0 0
38 41 1 0 0 0 0
30 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
S SKP 8
ID FL2FAAGS0013
KNApSAcK_ID C00008213
NAME Prunin 3'',6''-di-p-coumarate
CAS_RN 84823-66-5
FORMULA C39H34O14
EXACTMASS 726.194855796
AVERAGEMASS 726.67886
SMILES O(C(C2O)C(C(COC(C=Cc(c6)ccc(c6)O)=O)OC(Oc(c5)cc(c(c5O)4)OC(CC4=O)c(c3)ccc(c3)O)2)O)C(=O)C=Cc(c1)ccc(O)c1
M END