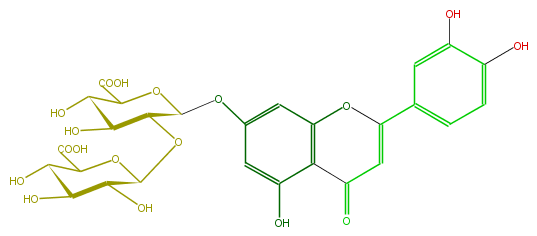
Mol:FL3FACGS0028
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.4731 -0.1393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4731 -0.1393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4731 -0.9489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4731 -0.9489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2279 -1.3536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2279 -1.3536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9290 -0.9489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9290 -0.9489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9290 -0.1393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9290 -0.1393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2279 0.2654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2279 0.2654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6301 -1.3536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6301 -1.3536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3311 -0.9489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3311 -0.9489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3311 -0.1393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3311 -0.1393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6301 0.2654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6301 0.2654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6302 -2.1124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6302 -2.1124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0318 0.2653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0318 0.2653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7465 -0.1472 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7465 -0.1472 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4610 0.2653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4610 0.2653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4610 1.0902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4610 1.0902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7465 1.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7465 1.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0318 1.0902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0318 1.0902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2279 -2.1613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2279 -2.1613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1591 1.4985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1591 1.4985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7479 2.1613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7479 2.1613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0087 0.3961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0087 0.3961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5399 -0.9732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5399 -0.9732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0435 -1.6284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0435 -1.6284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3287 -1.3505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3287 -1.3505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6389 -1.3429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6389 -1.3429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1401 -0.8416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1401 -0.8416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8703 -1.1038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8703 -1.1038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0949 -1.2936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0949 -1.2936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8174 -1.6660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8174 -1.6660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6083 -1.8537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6083 -1.8537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6943 0.4341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6943 0.4341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1979 -0.2212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1979 -0.2212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4830 0.0568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4830 0.0568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7932 0.0643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7932 0.0643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2944 0.5656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2944 0.5656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0247 0.3034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0247 0.3034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2493 0.1137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2493 0.1137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9717 -0.2588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9717 -0.2588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8451 -0.4877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8451 -0.4877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5193 0.7312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5193 0.7312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3135 0.7312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3135 0.7312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1208 1.4215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1208 1.4215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3650 -0.6348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3650 -0.6348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1591 -0.6348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1591 -0.6348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9664 0.0555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9664 0.0555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 21 34 1 0 0 0 0 | + | 21 34 1 0 0 0 0 |
| − | 39 25 1 0 0 0 0 | + | 39 25 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 40 42 1 0 0 0 0 | + | 40 42 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 27 43 1 0 0 0 0 | + | 27 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 40 41 42 | + | M SAL 1 3 40 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^ COOH | + | M SMT 1 ^ COOH |
| − | M SBV 1 46 0.4947 -0.4278 | + | M SBV 1 46 0.4947 -0.4278 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 43 44 45 | + | M SAL 2 3 43 44 45 |
| − | M SBL 2 1 49 | + | M SBL 2 1 49 |
| − | M SMT 2 ^ COOH | + | M SMT 2 ^ COOH |
| − | M SBV 2 49 0.4947 -0.4690 | + | M SBV 2 49 0.4947 -0.4690 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FACGS0028 | + | ID FL3FACGS0028 |
| − | FORMULA C27H26O18 | + | FORMULA C27H26O18 |
| − | EXACTMASS 638.111914028 | + | EXACTMASS 638.111914028 |
| − | AVERAGEMASS 638.4845399999999 | + | AVERAGEMASS 638.4845399999999 |
| − | SMILES O(C=4c(c5)cc(c(c5)O)O)c(c(C(=O)C4)3)cc(cc3O)OC(O1)C(OC(O2)C(O)C(C(C2C(O)=O)O)O)C(O)C(C1C(O)=O)O | + | SMILES O(C=4c(c5)cc(c(c5)O)O)c(c(C(=O)C4)3)cc(cc3O)OC(O1)C(OC(O2)C(O)C(C(C2C(O)=O)O)O)C(O)C(C1C(O)=O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-0.4731 -0.1393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4731 -0.9489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2279 -1.3536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9290 -0.9489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9290 -0.1393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2279 0.2654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6301 -1.3536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3311 -0.9489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3311 -0.1393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6301 0.2654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6302 -2.1124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0318 0.2653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7465 -0.1472 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4610 0.2653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4610 1.0902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7465 1.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0318 1.0902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2279 -2.1613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1591 1.4985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7479 2.1613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0087 0.3961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5399 -0.9732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0435 -1.6284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3287 -1.3505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6389 -1.3429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1401 -0.8416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8703 -1.1038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0949 -1.2936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8174 -1.6660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6083 -1.8537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6943 0.4341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1979 -0.2212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4830 0.0568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7932 0.0643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2944 0.5656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0247 0.3034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2493 0.1137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9717 -0.2588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8451 -0.4877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5193 0.7312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3135 0.7312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1208 1.4215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3650 -0.6348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1591 -0.6348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9664 0.0555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
15 19 1 0 0 0 0
16 20 1 0 0 0 0
1 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
21 34 1 0 0 0 0
39 25 1 0 0 0 0
40 41 2 0 0 0 0
40 42 1 0 0 0 0
36 40 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
27 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 40 41 42
M SBL 1 1 46
M SMT 1 ^ COOH
M SBV 1 46 0.4947 -0.4278
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 43 44 45
M SBL 2 1 49
M SMT 2 ^ COOH
M SBV 2 49 0.4947 -0.4690
S SKP 5
ID FL3FACGS0028
FORMULA C27H26O18
EXACTMASS 638.111914028
AVERAGEMASS 638.4845399999999
SMILES O(C=4c(c5)cc(c(c5)O)O)c(c(C(=O)C4)3)cc(cc3O)OC(O1)C(OC(O2)C(O)C(C(C2C(O)=O)O)O)C(O)C(C1C(O)=O)O
M END