Mol:FL3FACGS0039
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.4568 -1.1559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4568 -1.1559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4568 -1.6767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4568 -1.6767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0057 -1.9371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0057 -1.9371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5546 -1.6767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5546 -1.6767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5546 -1.1559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5546 -1.1559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0057 -0.8954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0057 -0.8954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1036 -1.9371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1036 -1.9371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6525 -1.6767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6525 -1.6767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6525 -1.1559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6525 -1.1559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1036 -0.8954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1036 -0.8954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1036 -2.3432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1036 -2.3432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2016 -0.8955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2016 -0.8955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7419 -1.1609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7419 -1.1609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2822 -0.8955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2822 -0.8955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2822 -0.3647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2822 -0.3647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7419 -0.0993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7419 -0.0993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2016 -0.3647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2016 -0.3647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9763 -0.8559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9763 -0.8559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0057 -2.4569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0057 -2.4569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3122 -0.0216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3122 -0.0216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7419 0.4315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7419 0.4315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6418 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6418 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2542 -0.4765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2542 -0.4765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9996 -0.2635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9996 -0.2635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7496 -0.4765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7496 -0.4765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1373 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1373 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3918 -0.0181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3918 -0.0181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9219 0.0020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9219 0.0020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6988 0.5136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6988 0.5136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6967 -0.1280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6967 -0.1280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1004 1.4078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1004 1.4078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4631 2.0360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4631 2.0360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7656 1.8366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7656 1.8366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0639 2.0360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0639 2.0360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7011 1.4078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7011 1.4078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3987 1.6070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3987 1.6070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3987 2.3774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3987 2.3774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7026 1.5691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7026 1.5691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0288 1.8844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0288 1.8844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1237 2.4569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1237 2.4569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2618 -0.4498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2618 -0.4498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9763 -0.8623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9763 -0.8623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 20 1 0 0 0 0 | + | 23 20 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 35 28 1 0 0 0 0 | + | 35 28 1 0 0 0 0 |
| − | 25 41 1 0 0 0 0 | + | 25 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -8.9175 3.5835 | + | M SBV 1 45 -8.9175 3.5835 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACGS0039 | + | ID FL3FACGS0039 |
| − | KNApSAcK_ID C00004299 | + | KNApSAcK_ID C00004299 |
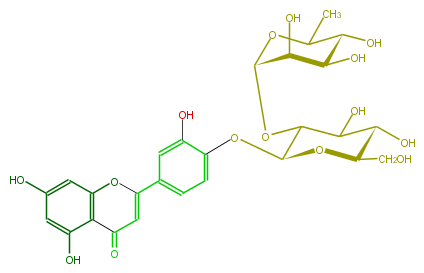
| − | NAME Luteolin 4'-neohesperidoside | + | NAME Luteolin 4'-neohesperidoside |
| − | CAS_RN 70404-48-7 | + | CAS_RN 70404-48-7 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES c(O)(c1)cc(c(C5=O)c1OC(=C5)c(c4)cc(O)c(c4)OC(O2)C(OC(O3)C(C(O)C(O)C(C)3)O)C(O)C(O)C2CO)O | + | SMILES c(O)(c1)cc(c(C5=O)c1OC(=C5)c(c4)cc(O)c(c4)OC(O2)C(OC(O3)C(C(O)C(O)C(C)3)O)C(O)C(O)C2CO)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-3.4568 -1.1559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4568 -1.6767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0057 -1.9371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5546 -1.6767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5546 -1.1559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0057 -0.8954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1036 -1.9371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6525 -1.6767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6525 -1.1559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1036 -0.8954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1036 -2.3432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2016 -0.8955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7419 -1.1609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2822 -0.8955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2822 -0.3647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7419 -0.0993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2016 -0.3647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9763 -0.8559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0057 -2.4569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3122 -0.0216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7419 0.4315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6418 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2542 -0.4765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9996 -0.2635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7496 -0.4765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1373 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3918 -0.0181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9219 0.0020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6988 0.5136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6967 -0.1280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1004 1.4078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4631 2.0360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7656 1.8366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0639 2.0360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7011 1.4078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3987 1.6070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3987 2.3774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7026 1.5691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0288 1.8844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1237 2.4569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2618 -0.4498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9763 -0.8623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 1 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 20 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
36 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
33 40 1 0 0 0 0
35 28 1 0 0 0 0
25 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -8.9175 3.5835
S SKP 8
ID FL3FACGS0039
KNApSAcK_ID C00004299
NAME Luteolin 4'-neohesperidoside
CAS_RN 70404-48-7
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES c(O)(c1)cc(c(C5=O)c1OC(=C5)c(c4)cc(O)c(c4)OC(O2)C(OC(O3)C(C(O)C(O)C(C)3)O)C(O)C(O)C2CO)O
M END