Mol:FL3FFCGS0016
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 35 38 0 0 0 0 0 0 0 0999 V2000 | + | 35 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.6967 -1.3063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6967 -1.3063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6967 -2.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6967 -2.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9956 -2.5848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9956 -2.5848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2945 -2.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2945 -2.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2945 -1.3705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2945 -1.3705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9956 -0.9656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9956 -0.9656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5933 -2.5848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5933 -2.5848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1076 -2.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1076 -2.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1076 -1.3705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1076 -1.3705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5933 -0.9656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5933 -0.9656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5933 -3.3323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5933 -3.3323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8085 -0.9658 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8085 -0.9658 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5232 -1.3783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5232 -1.3783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2377 -0.9658 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2377 -0.9658 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2377 -0.1407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2377 -0.1407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5232 0.2718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5232 0.2718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8085 -0.1407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8085 -0.1407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9956 -3.2696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9956 -3.2696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4007 -0.8417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4007 -0.8417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9908 -0.2125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9908 -0.2125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5232 1.0714 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5232 1.0714 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1722 2.7191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1722 2.7191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7483 1.8271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7483 1.8271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0318 1.1841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0318 1.1841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7813 0.2485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7813 0.2485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3943 1.0783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3943 1.0783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0350 1.8486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0350 1.8486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8049 3.3323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8049 3.3323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0519 2.6040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0519 2.6040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3126 0.1549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3126 0.1549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6467 2.5740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6467 2.5740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1339 1.6856 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1339 1.6856 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6764 2.5740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6764 2.5740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9413 0.2655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9413 0.2655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0519 -0.3756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0519 -0.3756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 6 20 1 0 0 0 0 | + | 6 20 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 31 32 2 0 0 0 0 | + | 31 32 2 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 15 34 1 0 0 0 0 | + | 15 34 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 31 32 33 | + | M SAL 1 3 31 32 33 |
| − | M SBL 1 1 36 | + | M SBL 1 1 36 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SBV 1 36 -0.3883 -0.7254 | + | M SBV 1 36 -0.3883 -0.7254 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 34 35 | + | M SAL 2 2 34 35 |
| − | M SBL 2 1 38 | + | M SBL 2 1 38 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 38 -0.7037 -0.4062 | + | M SBV 2 38 -0.7037 -0.4062 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FFCGS0016 | + | ID FL3FFCGS0016 |
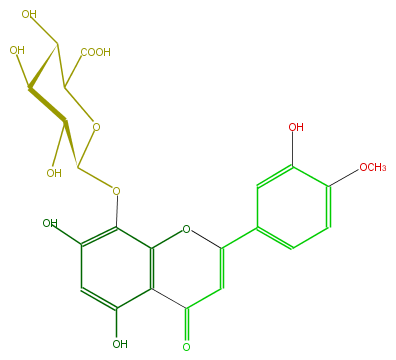
| − | FORMULA C22H20O13 | + | FORMULA C22H20O13 |
| − | EXACTMASS 492.090390726 | + | EXACTMASS 492.090390726 |
| − | AVERAGEMASS 492.3864 | + | AVERAGEMASS 492.3864 |
| − | SMILES Oc(c24)cc(c(c(OC(=CC4=O)c(c3)ccc(c3O)OC)2)OC(C(O)1)OC(C(O)C1O)C(O)=O)O | + | SMILES Oc(c24)cc(c(c(OC(=CC4=O)c(c3)ccc(c3O)OC)2)OC(C(O)1)OC(C(O)C1O)C(O)=O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
35 38 0 0 0 0 0 0 0 0999 V2000
-2.6967 -1.3063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6967 -2.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9956 -2.5848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2945 -2.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2945 -1.3705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9956 -0.9656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5933 -2.5848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1076 -2.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1076 -1.3705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5933 -0.9656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5933 -3.3323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8085 -0.9658 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5232 -1.3783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2377 -0.9658 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2377 -0.1407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5232 0.2718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8085 -0.1407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9956 -3.2696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4007 -0.8417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9908 -0.2125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5232 1.0714 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1722 2.7191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7483 1.8271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0318 1.1841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7813 0.2485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3943 1.0783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0350 1.8486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8049 3.3323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0519 2.6040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3126 0.1549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6467 2.5740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1339 1.6856 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6764 2.5740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9413 0.2655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0519 -0.3756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
6 20 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 20 1 0 0 0 0
31 32 2 0 0 0 0
31 33 1 0 0 0 0
27 31 1 0 0 0 0
34 35 1 0 0 0 0
15 34 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 31 32 33
M SBL 1 1 36
M SMT 1 COOH
M SBV 1 36 -0.3883 -0.7254
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 34 35
M SBL 2 1 38
M SMT 2 OCH3
M SBV 2 38 -0.7037 -0.4062
S SKP 5
ID FL3FFCGS0016
FORMULA C22H20O13
EXACTMASS 492.090390726
AVERAGEMASS 492.3864
SMILES Oc(c24)cc(c(c(OC(=CC4=O)c(c3)ccc(c3O)OC)2)OC(C(O)1)OC(C(O)C1O)C(O)=O)O
M END