Mol:FL5FACGSS001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 38 41 0 0 0 0 0 0 0 0999 V2000 | + | 38 41 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.4746 0.7893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4746 0.7893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4746 -0.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4746 -0.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7734 -0.4253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7734 -0.4253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0723 -0.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0723 -0.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0722 0.7893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0722 0.7893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7734 1.1941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7734 1.1941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3710 -0.4253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3710 -0.4253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3301 -0.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3301 -0.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3302 0.7893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3302 0.7893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3711 1.1941 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3711 1.1941 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3710 -1.1076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3710 -1.1076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0311 1.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0311 1.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7457 0.7813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7457 0.7813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4605 1.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4605 1.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4604 2.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4604 2.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7457 2.4317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7457 2.4317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0311 2.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0311 2.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1066 -0.4661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1066 -0.4661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1531 -0.6520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1531 -0.6520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4185 -0.9194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4185 -0.9194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1325 -1.2380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1325 -1.2380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5737 -1.8887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5737 -1.8887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3083 -1.6214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3083 -1.6214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5944 -1.3027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5944 -1.3027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5275 -0.2908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5275 -0.2908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4353 -1.2217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4353 -1.2217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5197 -2.0371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5197 -2.0371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1748 2.4316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1748 2.4316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7734 -1.0665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7734 -1.0665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3595 1.3871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3595 1.3871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6932 1.3871 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6932 1.3871 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9945 1.3870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9945 1.3870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6933 2.0117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6933 2.0117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6932 0.8146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6932 0.8146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7458 3.2567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7458 3.2567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4119 -2.4374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4119 -2.4374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3595 -2.0926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3595 -2.0926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2675 -3.2567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2675 -3.2567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 31 33 2 0 0 0 0 | + | 31 33 2 0 0 0 0 |
| − | 31 34 2 0 0 0 0 | + | 31 34 2 0 0 0 0 |
| − | 1 32 1 0 0 0 0 | + | 1 32 1 0 0 0 0 |
| − | 16 35 1 0 0 0 0 | + | 16 35 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 22 36 1 0 0 0 0 | + | 22 36 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 36 37 38 | + | M SAL 1 3 36 37 38 |
| − | M SBL 1 1 41 | + | M SBL 1 1 41 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SBV 1 41 -0.8383 0.5488 | + | M SBV 1 41 -0.8383 0.5488 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGSS001 | + | ID FL5FACGSS001 |
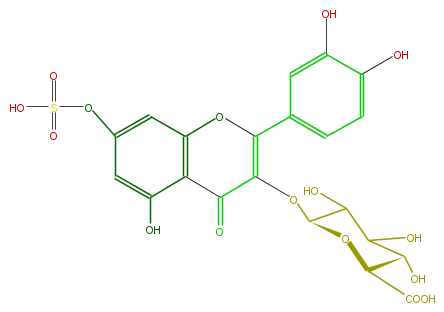
| − | FORMULA C21H18O16S | + | FORMULA C21H18O16S |
| − | EXACTMASS 558.031555218 | + | EXACTMASS 558.031555218 |
| − | AVERAGEMASS 558.42402 | + | AVERAGEMASS 558.42402 |
| − | SMILES C(C(C(O)=O)1)(O)C(C(C(OC(C4=O)=C(Oc(c34)cc(cc3O)OS(O)(=O)=O)c(c2)cc(c(O)c2)O)O1)O)O | + | SMILES C(C(C(O)=O)1)(O)C(C(C(OC(C4=O)=C(Oc(c34)cc(cc3O)OS(O)(=O)=O)c(c2)cc(c(O)c2)O)O1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
38 41 0 0 0 0 0 0 0 0999 V2000
-2.4746 0.7893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4746 -0.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7734 -0.4253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0723 -0.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0722 0.7893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7734 1.1941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3710 -0.4253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3301 -0.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3302 0.7893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3711 1.1941 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3710 -1.1076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0311 1.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7457 0.7813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4605 1.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4604 2.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7457 2.4317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0311 2.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1066 -0.4661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1531 -0.6520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4185 -0.9194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1325 -1.2380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5737 -1.8887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3083 -1.6214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5944 -1.3027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5275 -0.2908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4353 -1.2217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5197 -2.0371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1748 2.4316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7734 -1.0665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3595 1.3871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6932 1.3871 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0
-2.9945 1.3870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6933 2.0117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6932 0.8146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7458 3.2567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4119 -2.4374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3595 -2.0926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2675 -3.2567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
31 33 2 0 0 0 0
31 34 2 0 0 0 0
1 32 1 0 0 0 0
16 35 1 0 0 0 0
36 37 2 0 0 0 0
36 38 1 0 0 0 0
22 36 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 36 37 38
M SBL 1 1 41
M SMT 1 COOH
M SBV 1 41 -0.8383 0.5488
S SKP 5
ID FL5FACGSS001
FORMULA C21H18O16S
EXACTMASS 558.031555218
AVERAGEMASS 558.42402
SMILES C(C(C(O)=O)1)(O)C(C(C(OC(C4=O)=C(Oc(c34)cc(cc3O)OS(O)(=O)=O)c(c2)cc(c(O)c2)O)O1)O)O
M END