Mol:FL5FADGA0013
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.8563 0.7805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8563 0.7805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8563 -0.0445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8563 -0.0445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1419 -0.4570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1419 -0.4570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4275 -0.0445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4275 -0.0445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4275 0.7805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4275 0.7805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1419 1.1930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1419 1.1930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7131 -0.4570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7131 -0.4570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9986 -0.0445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9986 -0.0445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9986 0.7805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9986 0.7805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7131 1.1930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7131 1.1930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7131 -1.1001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7131 -1.1001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2845 1.1929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2845 1.1929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4437 0.7724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4437 0.7724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1719 1.1929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1719 1.1929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1719 2.0336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1719 2.0336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4437 2.4540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4437 2.4540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2845 2.0336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2845 2.0336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5705 1.1929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5705 1.1929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2641 -0.5247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2641 -0.5247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1419 -1.2816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1419 -1.2816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8318 2.5298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8318 2.5298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1742 -1.5668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1742 -1.5668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5742 -1.1347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5742 -1.1347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3368 -1.9657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3368 -1.9657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5742 -2.8017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5742 -2.8017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1742 -3.2339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1742 -3.2339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0632 -2.4029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0632 -2.4029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0430 -0.8482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0430 -0.8482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4277 -2.7877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4277 -2.7877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9158 -2.9873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9158 -2.9873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3590 -3.5227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3590 -3.5227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0488 -0.4102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0488 -0.4102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6169 -1.1601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6169 -1.1601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4350 -0.8420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4350 -0.8420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2245 -0.8335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2245 -0.8335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6509 -0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6509 -0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9350 -0.6374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9350 -0.6374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7532 -1.0809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7532 -1.0809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1115 -1.3003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1115 -1.3003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8389 -0.5303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8389 -0.5303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2299 -4.0168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2299 -4.0168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5546 -4.4067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5546 -4.4067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7688 -3.6570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7688 -3.6570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5546 -2.9028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5546 -2.9028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2299 -2.5127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2299 -2.5127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0156 -3.2626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0156 -3.2626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1705 -1.9997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1705 -1.9997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7284 -2.2843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7284 -2.2843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6217 -2.7711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6217 -2.7711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7783 -4.0267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7783 -4.0267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8473 -4.0054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8473 -4.0054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3577 -0.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3577 -0.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5705 -0.4850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5705 -0.4850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4584 3.2544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4584 3.2544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0253 4.4067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0253 4.4067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 28 32 1 0 0 0 0 | + | 28 32 1 0 0 0 0 |
| − | 35 40 1 0 0 0 0 | + | 35 40 1 0 0 0 0 |
| − | 42 41 1 1 0 0 0 | + | 42 41 1 1 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 46 41 1 1 0 0 0 | + | 46 41 1 1 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 44 48 1 0 0 0 0 | + | 44 48 1 0 0 0 0 |
| − | 46 49 1 0 0 0 0 | + | 46 49 1 0 0 0 0 |
| − | 41 50 1 0 0 0 0 | + | 41 50 1 0 0 0 0 |
| − | 42 51 1 0 0 0 0 | + | 42 51 1 0 0 0 0 |
| − | 51 31 1 0 0 0 0 | + | 51 31 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 36 52 1 0 0 0 0 | + | 36 52 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 16 54 1 0 0 0 0 | + | 16 54 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 52 53 | + | M SAL 1 2 52 53 |
| − | M SBL 1 1 58 | + | M SBL 1 1 58 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 58 -0.7068 -0.1971 | + | M SBV 1 58 -0.7068 -0.1971 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 54 55 | + | M SAL 2 2 54 55 |
| − | M SBL 2 1 60 | + | M SBL 2 1 60 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 60 -0.0147 -0.8004 | + | M SBV 2 60 -0.0147 -0.8004 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGA0013 | + | ID FL5FADGA0013 |
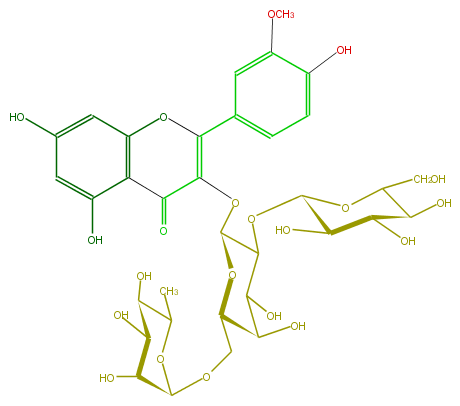
| − | FORMULA C34H42O21 | + | FORMULA C34H42O21 |
| − | EXACTMASS 786.2218584059999 | + | EXACTMASS 786.2218584059999 |
| − | AVERAGEMASS 786.68468 | + | AVERAGEMASS 786.68468 |
| − | SMILES C(C6O)(C(O)C(OC(C)6)OCC(O1)C(O)C(O)C(OC(O5)C(O)C(C(O)C5CO)O)C(OC(C3=O)=C(c(c4)cc(OC)c(c4)O)Oc(c32)cc(O)cc(O)2)1)O | + | SMILES C(C6O)(C(O)C(OC(C)6)OCC(O1)C(O)C(O)C(OC(O5)C(O)C(C(O)C5CO)O)C(OC(C3=O)=C(c(c4)cc(OC)c(c4)O)Oc(c32)cc(O)cc(O)2)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-3.8563 0.7805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8563 -0.0445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1419 -0.4570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4275 -0.0445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4275 0.7805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1419 1.1930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7131 -0.4570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9986 -0.0445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9986 0.7805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7131 1.1930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7131 -1.1001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2845 1.1929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4437 0.7724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1719 1.1929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1719 2.0336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4437 2.4540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2845 2.0336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5705 1.1929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2641 -0.5247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1419 -1.2816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8318 2.5298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1742 -1.5668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5742 -1.1347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3368 -1.9657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5742 -2.8017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1742 -3.2339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0632 -2.4029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0430 -0.8482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4277 -2.7877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9158 -2.9873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3590 -3.5227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0488 -0.4102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6169 -1.1601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4350 -0.8420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2245 -0.8335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6509 -0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9350 -0.6374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7532 -1.0809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1115 -1.3003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8389 -0.5303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2299 -4.0168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5546 -4.4067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7688 -3.6570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5546 -2.9028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2299 -2.5127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0156 -3.2626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1705 -1.9997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7284 -2.2843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6217 -2.7711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7783 -4.0267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8473 -4.0054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3577 -0.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5705 -0.4850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4584 3.2544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0253 4.4067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 15 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
25 31 1 0 0 0 0
23 19 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
33 38 1 0 0 0 0
34 39 1 0 0 0 0
28 32 1 0 0 0 0
35 40 1 0 0 0 0
42 41 1 1 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 46 1 1 0 0 0
46 41 1 1 0 0 0
45 47 1 0 0 0 0
44 48 1 0 0 0 0
46 49 1 0 0 0 0
41 50 1 0 0 0 0
42 51 1 0 0 0 0
51 31 1 0 0 0 0
52 53 1 0 0 0 0
36 52 1 0 0 0 0
54 55 1 0 0 0 0
16 54 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 52 53
M SBL 1 1 58
M SMT 1 CH2OH
M SBV 1 58 -0.7068 -0.1971
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 54 55
M SBL 2 1 60
M SMT 2 OCH3
M SBV 2 60 -0.0147 -0.8004
S SKP 5
ID FL5FADGA0013
FORMULA C34H42O21
EXACTMASS 786.2218584059999
AVERAGEMASS 786.68468
SMILES C(C6O)(C(O)C(OC(C)6)OCC(O1)C(O)C(O)C(OC(O5)C(O)C(C(O)C5CO)O)C(OC(C3=O)=C(c(c4)cc(OC)c(c4)O)Oc(c32)cc(O)cc(O)2)1)O
M END