Mol:FL5FBCGL0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.5444 1.0319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5444 1.0319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5444 0.2225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5444 0.2225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8434 -0.1823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8434 -0.1823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1423 0.2225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1423 0.2225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1423 1.0319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1423 1.0319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8434 1.4366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8434 1.4366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4413 -0.1823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4413 -0.1823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7403 0.2225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7403 0.2225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7403 1.0319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7403 1.0319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4413 1.4366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4413 1.4366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4413 -0.8134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4413 -0.8134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0395 1.4365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0395 1.4365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6750 1.0240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6750 1.0240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3895 1.4365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3895 1.4365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3895 2.2615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3895 2.2615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6750 2.6740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6750 2.6740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0395 2.2615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0395 2.2615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3320 -1.8284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3320 -1.8284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8435 -2.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8435 -2.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7829 -2.4059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7829 -2.4059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7280 -2.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7280 -2.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2451 -1.8487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2451 -1.8487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2772 -2.0968 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2772 -2.0968 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5292 -1.9904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5292 -1.9904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3472 0.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3472 0.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8587 -0.6063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8587 -0.6063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7982 -0.3378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7982 -0.3378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7432 -0.6063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7432 -0.6063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2318 0.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2318 0.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2924 -0.0285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2924 -0.0285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0812 -0.2949 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0812 -0.2949 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9280 0.5987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9280 0.5987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0010 -0.0115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0010 -0.0115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5577 -0.4839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5577 -0.4839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1445 -1.0228 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1445 -1.0228 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2343 -2.5113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2343 -2.5113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1911 -3.0404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1911 -3.0404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7280 -3.4987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7280 -3.4987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5220 0.0940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5220 0.0940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2451 1.4365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2451 1.4365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1037 2.6738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1037 2.6738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6750 3.4987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6750 3.4987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8434 -0.8879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8434 -0.8879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9430 -1.4077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9430 -1.4077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 18 1 0 0 0 0 | + | 23 18 1 0 0 0 0 |
| − | 18 24 1 0 0 0 0 | + | 18 24 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 29 28 1 1 0 0 0 | + | 29 28 1 1 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 25 1 0 0 0 0 | + | 30 25 1 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 29 33 1 0 0 0 0 | + | 29 33 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 22 35 1 0 0 0 0 | + | 22 35 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 19 36 1 0 0 0 0 | + | 19 36 1 0 0 0 0 |
| − | 20 37 1 0 0 0 0 | + | 20 37 1 0 0 0 0 |
| − | 21 38 1 0 0 0 0 | + | 21 38 1 0 0 0 0 |
| − | 31 8 1 0 0 0 0 | + | 31 8 1 0 0 0 0 |
| − | 26 31 1 0 0 0 0 | + | 26 31 1 0 0 0 0 |
| − | 25 39 1 0 0 0 0 | + | 25 39 1 0 0 0 0 |
| − | 1 40 1 0 0 0 0 | + | 1 40 1 0 0 0 0 |
| − | 15 41 1 0 0 0 0 | + | 15 41 1 0 0 0 0 |
| − | 16 42 1 0 0 0 0 | + | 16 42 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 3 43 1 0 0 0 0 | + | 3 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 48 | + | M SBL 1 1 48 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 48 0.0000 0.7056 | + | M SBV 1 48 0.0000 0.7056 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FBCGL0002 | + | ID FL5FBCGL0002 |
| − | FORMULA C28H32O16 | + | FORMULA C28H32O16 |
| − | EXACTMASS 624.1690349759999 | + | EXACTMASS 624.1690349759999 |
| − | AVERAGEMASS 624.54408 | + | AVERAGEMASS 624.54408 |
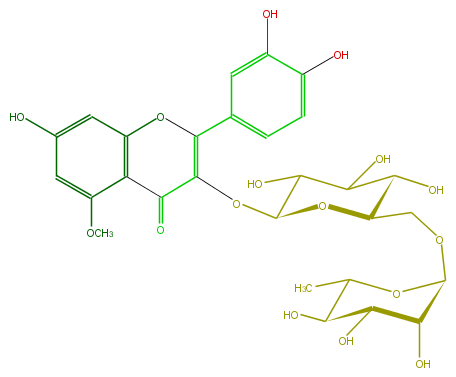
| − | SMILES C(C1COC(O5)C(C(C(O)C(C)5)O)O)(C(O)C(O)C(OC(C4=O)=C(Oc(c34)cc(O)cc3OC)c(c2)ccc(c2O)O)O1)O | + | SMILES C(C1COC(O5)C(C(C(O)C(C)5)O)O)(C(O)C(O)C(OC(C4=O)=C(Oc(c34)cc(O)cc3OC)c(c2)ccc(c2O)O)O1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-3.5444 1.0319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5444 0.2225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8434 -0.1823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1423 0.2225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1423 1.0319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8434 1.4366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4413 -0.1823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7403 0.2225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7403 1.0319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4413 1.4366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4413 -0.8134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0395 1.4365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6750 1.0240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3895 1.4365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3895 2.2615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6750 2.6740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0395 2.2615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3320 -1.8284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8435 -2.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7829 -2.4059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7280 -2.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2451 -1.8487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2772 -2.0968 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5292 -1.9904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3472 0.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8587 -0.6063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7982 -0.3378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7432 -0.6063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2318 0.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2924 -0.0285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0812 -0.2949 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9280 0.5987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0010 -0.0115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5577 -0.4839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1445 -1.0228 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2343 -2.5113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1911 -3.0404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7280 -3.4987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5220 0.0940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2451 1.4365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1037 2.6738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6750 3.4987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8434 -0.8879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9430 -1.4077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 19 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 18 1 0 0 0 0
18 24 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 28 1 1 0 0 0
29 28 1 1 0 0 0
29 30 1 0 0 0 0
30 25 1 0 0 0 0
30 32 1 0 0 0 0
29 33 1 0 0 0 0
28 34 1 0 0 0 0
22 35 1 0 0 0 0
34 35 1 0 0 0 0
19 36 1 0 0 0 0
20 37 1 0 0 0 0
21 38 1 0 0 0 0
31 8 1 0 0 0 0
26 31 1 0 0 0 0
25 39 1 0 0 0 0
1 40 1 0 0 0 0
15 41 1 0 0 0 0
16 42 1 0 0 0 0
43 44 1 0 0 0 0
3 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 48
M SMT 1 OCH3
M SBV 1 48 0.0000 0.7056
S SKP 5
ID FL5FBCGL0002
FORMULA C28H32O16
EXACTMASS 624.1690349759999
AVERAGEMASS 624.54408
SMILES C(C1COC(O5)C(C(C(O)C(C)5)O)O)(C(O)C(O)C(OC(C4=O)=C(Oc(c34)cc(O)cc3OC)c(c2)ccc(c2O)O)O1)O
M END