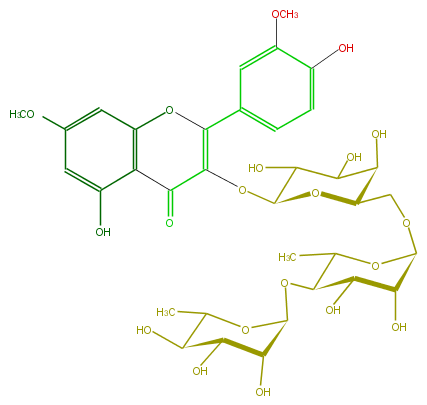
Mol:FL5FCDGA0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.9224 0.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9224 0.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9224 0.0674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9224 0.0674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2214 -0.3374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2214 -0.3374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5204 0.0674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5204 0.0674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5204 0.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5204 0.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2214 1.2815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2214 1.2815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8193 -0.3374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8193 -0.3374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1183 0.0674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1183 0.0674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1183 0.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1183 0.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8193 1.2815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8193 1.2815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8193 -0.9685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8193 -0.9685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5824 1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5824 1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2969 0.8689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2969 0.8689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0115 1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0115 1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0115 2.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0115 2.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2969 2.5189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2969 2.5189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5824 2.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5824 2.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6273 -0.3210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6273 -0.3210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2214 -1.1465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2214 -1.1465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4748 -1.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4748 -1.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0517 -2.3322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0517 -2.3322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8652 -2.0997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8652 -2.0997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6836 -2.3322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6836 -2.3322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1068 -1.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1068 -1.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2932 -1.8319 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2932 -1.8319 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6501 -1.6525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6501 -1.6525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6955 0.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6955 0.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2725 -0.6078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2725 -0.6078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0859 -0.3753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0859 -0.3753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9043 -0.6078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9043 -0.6078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3275 0.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3275 0.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5139 -0.1075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5139 -0.1075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9744 0.1179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9744 0.1179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7884 0.3475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7884 0.3475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2876 0.8059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2876 0.8059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5799 -0.4187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5799 -0.4187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9120 -0.9716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9120 -0.9716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4739 -2.1809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4739 -2.1809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3747 -2.7125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3747 -2.7125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6836 -3.0460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6836 -3.0460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6297 2.5289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6297 2.5289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1001 -2.8003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1001 -2.8003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5853 -3.4935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5853 -3.4935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2453 -3.3328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2453 -3.3328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0404 -3.6357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0404 -3.6357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5257 -2.9426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5257 -2.9426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6950 -3.1034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6950 -3.1034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7865 -2.7562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7865 -2.7562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2632 -3.1077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2632 -3.1077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2818 -3.9298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2818 -3.9298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9782 -4.3468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9782 -4.3468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2969 3.2161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2969 3.2161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8531 4.3468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8531 4.3468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4768 1.1969 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4768 1.1969 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1068 2.2883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1068 2.2883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 8 18 1 0 0 0 0 | + | 8 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 27 1 0 0 0 0 | + | 32 27 1 0 0 0 0 |
| − | 27 33 1 0 0 0 0 | + | 27 33 1 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 31 35 1 0 0 0 0 | + | 31 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 24 37 1 0 0 0 0 | + | 24 37 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 21 38 1 0 0 0 0 | + | 21 38 1 0 0 0 0 |
| − | 22 39 1 0 0 0 0 | + | 22 39 1 0 0 0 0 |
| − | 23 40 1 0 0 0 0 | + | 23 40 1 0 0 0 0 |
| − | 28 18 1 0 0 0 0 | + | 28 18 1 0 0 0 0 |
| − | 41 15 1 0 0 0 0 | + | 41 15 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 42 1 0 0 0 0 | + | 47 42 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 38 1 0 0 0 0 | + | 46 38 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 16 52 1 0 0 0 0 | + | 16 52 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 1 54 1 0 0 0 0 | + | 1 54 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 52 53 | + | M SAL 1 2 52 53 |
| − | M SBL 1 1 58 | + | M SBL 1 1 58 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 58 0.0000 -0.6973 | + | M SBV 1 58 0.0000 -0.6973 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 54 55 | + | M SAL 2 2 54 55 |
| − | M SBL 2 1 60 | + | M SBL 2 1 60 |
| − | M SMT 2 ^ OCH3 | + | M SMT 2 ^ OCH3 |
| − | M SBV 2 60 0.5544 -0.3201 | + | M SBV 2 60 0.5544 -0.3201 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FCDGA0004 | + | ID FL5FCDGA0004 |
| − | FORMULA C35H44O20 | + | FORMULA C35H44O20 |
| − | EXACTMASS 784.242593848 | + | EXACTMASS 784.242593848 |
| − | AVERAGEMASS 784.71186 | + | AVERAGEMASS 784.71186 |
| − | SMILES Oc(c6)c(c2cc(OC)6)C(C(OC(C(O)5)OC(C(O)C5O)COC(O4)C(O)C(O)C(C4C)OC(C3O)OC(C)C(C3O)O)=C(O2)c(c1)cc(c(c1)O)OC)=O | + | SMILES Oc(c6)c(c2cc(OC)6)C(C(OC(C(O)5)OC(C(O)C5O)COC(O4)C(O)C(O)C(C4C)OC(C3O)OC(C)C(C3O)O)=C(O2)c(c1)cc(c(c1)O)OC)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-2.9224 0.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9224 0.0674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2214 -0.3374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5204 0.0674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5204 0.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2214 1.2815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8193 -0.3374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1183 0.0674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1183 0.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8193 1.2815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8193 -0.9685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5824 1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2969 0.8689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0115 1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0115 2.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2969 2.5189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5824 2.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6273 -0.3210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2214 -1.1465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4748 -1.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0517 -2.3322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8652 -2.0997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6836 -2.3322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1068 -1.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2932 -1.8319 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6501 -1.6525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6955 0.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2725 -0.6078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0859 -0.3753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9043 -0.6078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3275 0.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5139 -0.1075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9744 0.1179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7884 0.3475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2876 0.8059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5799 -0.4187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9120 -0.9716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4739 -2.1809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3747 -2.7125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6836 -3.0460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6297 2.5289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1001 -2.8003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5853 -3.4935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2453 -3.3328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0404 -3.6357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5257 -2.9426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6950 -3.1034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7865 -2.7562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2632 -3.1077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2818 -3.9298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9782 -4.3468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2969 3.2161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8531 4.3468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4768 1.1969 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1068 2.2883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
8 18 1 0 0 0 0
3 19 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
27 28 1 0 0 0 0
28 29 1 1 0 0 0
29 30 1 1 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 27 1 0 0 0 0
27 33 1 0 0 0 0
32 34 1 0 0 0 0
31 35 1 0 0 0 0
30 36 1 0 0 0 0
24 37 1 0 0 0 0
36 37 1 0 0 0 0
21 38 1 0 0 0 0
22 39 1 0 0 0 0
23 40 1 0 0 0 0
28 18 1 0 0 0 0
41 15 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 42 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 38 1 0 0 0 0
52 53 1 0 0 0 0
16 52 1 0 0 0 0
54 55 1 0 0 0 0
1 54 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 52 53
M SBL 1 1 58
M SMT 1 OCH3
M SBV 1 58 0.0000 -0.6973
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 54 55
M SBL 2 1 60
M SMT 2 ^ OCH3
M SBV 2 60 0.5544 -0.3201
S SKP 5
ID FL5FCDGA0004
FORMULA C35H44O20
EXACTMASS 784.242593848
AVERAGEMASS 784.71186
SMILES Oc(c6)c(c2cc(OC)6)C(C(OC(C(O)5)OC(C(O)C5O)COC(O4)C(O)C(O)C(C4C)OC(C3O)OC(C)C(C3O)O)=C(O2)c(c1)cc(c(c1)O)OC)=O
M END