Mol:FL63AGNS0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.5243 0.6297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5243 0.6297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5243 0.0079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5243 0.0079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0143 -0.3030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0143 -0.3030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5528 0.0079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5528 0.0079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5528 0.6297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5528 0.6297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0143 0.9407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0143 0.9407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0913 -0.3030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0913 -0.3030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6298 0.0079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6298 0.0079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6298 0.6297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6298 0.6297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0913 0.9407 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0913 0.9407 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0625 0.9405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0625 0.9405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1094 0.9066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1094 0.9066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6541 0.5921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6541 0.5921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1988 0.9066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1988 0.9066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1988 1.5356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1988 1.5356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6541 1.8501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6541 1.8501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1094 1.5356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1094 1.5356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0143 -0.9245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0143 -0.9245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6541 2.4785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6541 2.4785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0518 -0.5420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0518 -0.5420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7430 1.8498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7430 1.8498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7430 0.5924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7430 0.5924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5515 -1.4903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5515 -1.4903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1783 -2.1367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1783 -2.1367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2466 -1.4903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2466 -1.4903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5336 -0.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5336 -0.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1076 -0.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1076 -0.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3947 -1.4903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3947 -1.4903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1076 -1.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1076 -1.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5336 -1.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5336 -1.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3943 -0.4966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3943 -0.4966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9680 -1.4903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9680 -1.4903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3943 -2.4840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3943 -2.4840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6007 0.6297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6007 0.6297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1390 0.9405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1390 0.9405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6007 0.0082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6007 0.0082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1390 1.5579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1390 1.5579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6737 1.8666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6737 1.8666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2083 1.5579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2083 1.5579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2083 0.9405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2083 0.9405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6737 0.6318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6737 0.6318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6737 2.4840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6737 2.4840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7430 1.8666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7430 1.8666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7430 0.6318 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7430 0.6318 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 1 11 1 0 0 0 0 | + | 1 11 1 0 0 0 0 |
| − | 9 12 1 6 0 0 0 | + | 9 12 1 6 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 8 20 1 6 0 0 0 | + | 8 20 1 6 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 14 22 1 0 0 0 0 | + | 14 22 1 0 0 0 0 |
| − | 18 23 1 0 0 0 0 | + | 18 23 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 2 0 0 0 0 | + | 29 30 2 0 0 0 0 |
| − | 30 25 1 0 0 0 0 | + | 30 25 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 28 32 1 0 0 0 0 | + | 28 32 1 0 0 0 0 |
| − | 29 33 1 0 0 0 0 | + | 29 33 1 0 0 0 0 |
| − | 11 34 1 0 0 0 0 | + | 11 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 34 36 2 0 0 0 0 | + | 34 36 2 0 0 0 0 |
| − | 35 37 2 0 0 0 0 | + | 35 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 35 1 0 0 0 0 | + | 41 35 1 0 0 0 0 |
| − | 38 42 1 0 0 0 0 | + | 38 42 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | 40 44 1 0 0 0 0 | + | 40 44 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL63AGNS0012 | + | ID FL63AGNS0012 |
| − | KNApSAcK_ID C00008890 | + | KNApSAcK_ID C00008890 |
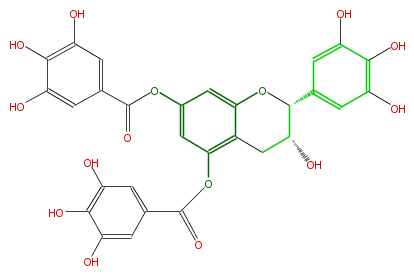
| − | NAME Epigallocatechin 5,7,-di-O-gallate | + | NAME Epigallocatechin 5,7,-di-O-gallate |
| − | CAS_RN 96658-19-4 | + | CAS_RN 96658-19-4 |
| − | FORMULA C29H22O15 | + | FORMULA C29H22O15 |
| − | EXACTMASS 610.095870034 | + | EXACTMASS 610.095870034 |
| − | AVERAGEMASS 610.4759799999999 | + | AVERAGEMASS 610.4759799999999 |
| − | SMILES O=C(Oc(c2)cc(OC(=O)c(c5)cc(c(c(O)5)O)O)c(C4)c(OC(C(O)4)c(c3)cc(c(c(O)3)O)O)2)c(c1)cc(c(c1O)O)O | + | SMILES O=C(Oc(c2)cc(OC(=O)c(c5)cc(c(c(O)5)O)O)c(C4)c(OC(C(O)4)c(c3)cc(c(c(O)3)O)O)2)c(c1)cc(c(c1O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-0.5243 0.6297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5243 0.0079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0143 -0.3030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5528 0.0079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5528 0.6297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0143 0.9407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0913 -0.3030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6298 0.0079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6298 0.6297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0913 0.9407 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0625 0.9405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1094 0.9066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6541 0.5921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1988 0.9066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1988 1.5356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6541 1.8501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1094 1.5356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0143 -0.9245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6541 2.4785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0518 -0.5420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7430 1.8498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7430 0.5924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5515 -1.4903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1783 -2.1367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2466 -1.4903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5336 -0.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1076 -0.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3947 -1.4903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1076 -1.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5336 -1.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3943 -0.4966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9680 -1.4903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3943 -2.4840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6007 0.6297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1390 0.9405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6007 0.0082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1390 1.5579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6737 1.8666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2083 1.5579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2083 0.9405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6737 0.6318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6737 2.4840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7430 1.8666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7430 0.6318 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
1 11 1 0 0 0 0
9 12 1 6 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
16 19 1 0 0 0 0
8 20 1 6 0 0 0
15 21 1 0 0 0 0
14 22 1 0 0 0 0
18 23 1 0 0 0 0
23 24 2 0 0 0 0
23 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 29 1 0 0 0 0
29 30 2 0 0 0 0
30 25 1 0 0 0 0
27 31 1 0 0 0 0
28 32 1 0 0 0 0
29 33 1 0 0 0 0
11 34 1 0 0 0 0
34 35 1 0 0 0 0
34 36 2 0 0 0 0
35 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 35 1 0 0 0 0
38 42 1 0 0 0 0
39 43 1 0 0 0 0
40 44 1 0 0 0 0
S SKP 8
ID FL63AGNS0012
KNApSAcK_ID C00008890
NAME Epigallocatechin 5,7,-di-O-gallate
CAS_RN 96658-19-4
FORMULA C29H22O15
EXACTMASS 610.095870034
AVERAGEMASS 610.4759799999999
SMILES O=C(Oc(c2)cc(OC(=O)c(c5)cc(c(c(O)5)O)O)c(C4)c(OC(C(O)4)c(c3)cc(c(c(O)3)O)O)2)c(c1)cc(c(c1O)O)O
M END