Mol:FL7AAGGL0010
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.2157 1.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2157 1.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2157 0.2845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2157 0.2845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5012 -0.1281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5012 -0.1281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7867 0.2845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7867 0.2845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7867 1.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7867 1.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5012 1.5219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5012 1.5219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0723 -0.1281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0723 -0.1281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6422 0.2845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6422 0.2845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6422 1.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6422 1.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0723 1.5219 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -0.0723 1.5219 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3564 1.5218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3564 1.5218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0846 1.1014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0846 1.1014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8127 1.5218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8127 1.5218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8127 2.3626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8127 2.3626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0846 2.7830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0846 2.7830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3564 2.3626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3564 2.3626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0846 3.6235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0846 3.6235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9299 1.5218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9299 1.5218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5406 2.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5406 2.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5012 -0.9527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5012 -0.9527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5406 1.1015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5406 1.1015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3674 -0.2257 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3674 -0.2257 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7499 -1.4095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7499 -1.4095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0987 -0.9116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0987 -0.9116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2671 -1.7863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2671 -1.7863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0427 -2.5760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0427 -2.5760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7499 -2.9845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7499 -2.9845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5255 -2.1994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5255 -2.1994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7681 -0.5801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7681 -0.5801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1297 -2.6901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1297 -2.6901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6100 -3.6235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6100 -3.6235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6281 -0.4549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6281 -0.4549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0180 -1.2152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0180 -1.2152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7998 -1.0597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7998 -1.0597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5070 -1.1765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5070 -1.1765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0845 -0.5729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0845 -0.5729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3848 -0.7979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3848 -0.7979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4679 -1.0741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4679 -1.0741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3437 -1.6118 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3437 -1.6118 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0907 -0.9848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0907 -0.9848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8649 -0.9136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8649 -0.9136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4230 -1.4971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4230 -1.4971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7866 -1.2496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7866 -1.2496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1725 -1.2429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1725 -1.2429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6188 -0.7966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6188 -0.7966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2688 -1.0300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2688 -1.0300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4317 -1.0509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4317 -1.0509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0657 -1.4906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0657 -1.4906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1524 -1.7187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1524 -1.7187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0173 -3.2630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0173 -3.2630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2667 -3.2350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2667 -3.2350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8479 -0.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8479 -0.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0907 -0.1828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0907 -0.1828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 15 17 1 0 0 0 0 | + | 15 17 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 13 21 1 0 0 0 0 | + | 13 21 1 0 0 0 0 |
| − | 22 8 1 0 0 0 0 | + | 22 8 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 35 40 1 0 0 0 0 | + | 35 40 1 0 0 0 0 |
| − | 29 32 1 0 0 0 0 | + | 29 32 1 0 0 0 0 |
| − | 24 22 1 0 0 0 0 | + | 24 22 1 0 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 44 43 1 1 0 0 0 | + | 44 43 1 1 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 20 1 0 0 0 0 | + | 44 20 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 26 50 1 0 0 0 0 | + | 26 50 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 46 52 1 0 0 0 0 | + | 46 52 1 0 0 0 0 |
| − | M CHG 1 10 1 | + | M CHG 1 10 1 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 56 0.0254 0.6870 | + | M SBV 1 56 0.0254 0.6870 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 58 0.5791 -0.5235 | + | M SBV 2 58 0.5791 -0.5235 |
| − | S SKP 5 | + | S SKP 5 |
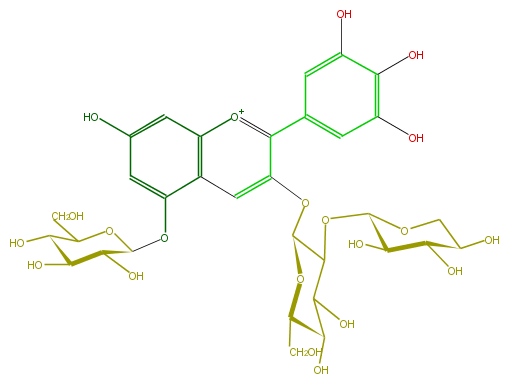
| − | ID FL7AAGGL0010 | + | ID FL7AAGGL0010 |
| − | FORMULA C32H39O21 | + | FORMULA C32H39O21 |
| − | EXACTMASS 759.19838331 | + | EXACTMASS 759.19838331 |
| − | AVERAGEMASS 759.63946 | + | AVERAGEMASS 759.63946 |
| − | SMILES c(c23)(cc(cc2[o+1]c(c(c6)cc(c(O)c(O)6)O)c(OC(C4OC(C5O)OCC(O)C5O)OC(CO)C(O)C4O)c3)O)OC(C(O)1)OC(CO)C(O)C1O | + | SMILES c(c23)(cc(cc2[o+1]c(c(c6)cc(c(O)c(O)6)O)c(OC(C4OC(C5O)OCC(O)C5O)OC(CO)C(O)C4O)c3)O)OC(C(O)1)OC(CO)C(O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-2.2157 1.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2157 0.2845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5012 -0.1281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7867 0.2845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7867 1.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5012 1.5219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0723 -0.1281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6422 0.2845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6422 1.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0723 1.5219 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.3564 1.5218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0846 1.1014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8127 1.5218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8127 2.3626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0846 2.7830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3564 2.3626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0846 3.6235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9299 1.5218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5406 2.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5012 -0.9527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5406 1.1015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3674 -0.2257 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7499 -1.4095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0987 -0.9116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2671 -1.7863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0427 -2.5760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7499 -2.9845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5255 -2.1994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7681 -0.5801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1297 -2.6901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6100 -3.6235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6281 -0.4549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0180 -1.2152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7998 -1.0597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5070 -1.1765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0845 -0.5729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3848 -0.7979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4679 -1.0741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3437 -1.6118 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0907 -0.9848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8649 -0.9136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4230 -1.4971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7866 -1.2496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1725 -1.2429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6188 -0.7966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2688 -1.0300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4317 -1.0509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0657 -1.4906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1524 -1.7187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0173 -3.2630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2667 -3.2350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8479 -0.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0907 -0.1828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
15 17 1 0 0 0 0
1 18 1 0 0 0 0
14 19 1 0 0 0 0
3 20 1 0 0 0 0
13 21 1 0 0 0 0
22 8 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
28 30 1 0 0 0 0
27 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
33 38 1 0 0 0 0
34 39 1 0 0 0 0
35 40 1 0 0 0 0
29 32 1 0 0 0 0
24 22 1 0 0 0 0
41 42 1 1 0 0 0
42 43 1 1 0 0 0
44 43 1 1 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
46 41 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 20 1 0 0 0 0
50 51 1 0 0 0 0
26 50 1 0 0 0 0
52 53 1 0 0 0 0
46 52 1 0 0 0 0
M CHG 1 10 1
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 56
M SMT 1 ^ CH2OH
M SBV 1 56 0.0254 0.6870
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 58
M SMT 2 ^ CH2OH
M SBV 2 58 0.5791 -0.5235
S SKP 5
ID FL7AAGGL0010
FORMULA C32H39O21
EXACTMASS 759.19838331
AVERAGEMASS 759.63946
SMILES c(c23)(cc(cc2[o+1]c(c(c6)cc(c(O)c(O)6)O)c(OC(C4OC(C5O)OCC(O)C5O)OC(CO)C(O)C4O)c3)O)OC(C(O)1)OC(CO)C(O)C1O
M END