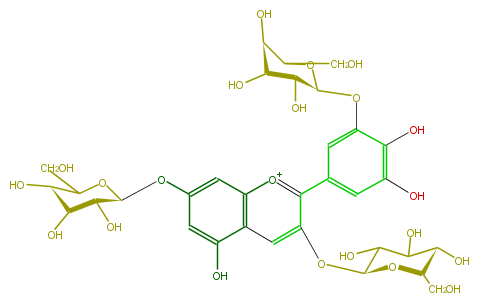
Mol:FL7AAGGL0014
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2169 -0.8609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2169 -0.8609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2169 -1.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2169 -1.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6606 -1.8244 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6606 -1.8244 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1043 -1.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1043 -1.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1043 -0.8609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1043 -0.8609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6606 -0.5397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6606 -0.5397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4520 -1.8244 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4520 -1.8244 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0083 -1.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0083 -1.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0083 -0.8609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0083 -0.8609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4520 -0.5397 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 0.4520 -0.5397 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5644 -0.5398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5644 -0.5398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1313 -0.8672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1313 -0.8672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6983 -0.5398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6983 -0.5398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6983 0.1149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6983 0.1149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1313 0.4422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1313 0.4422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5644 0.1149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5644 0.1149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7730 -0.5398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7730 -0.5398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2651 0.4421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2651 0.4421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6606 -2.4666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6606 -2.4666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4244 -2.2240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4244 -2.2240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1313 1.0967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1313 1.0967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2651 -0.8671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2651 -0.8671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5818 -1.9239 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.5818 -1.9239 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.2810 -2.4449 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.2810 -2.4449 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.8595 -2.2796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8595 -2.2796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4414 -2.4449 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.4414 -2.4449 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.7423 -1.9239 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.7423 -1.9239 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.1638 -2.0892 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.1638 -2.0892 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.0231 -2.0736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0231 -2.0736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2053 -1.6146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2053 -1.6146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1764 -2.1745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1764 -2.1745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9950 -0.6989 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.9950 -0.6989 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.6238 -1.1888 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.6238 -1.1888 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.0894 -0.9810 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.0894 -0.9810 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.5736 -0.9754 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.5736 -0.9754 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.9484 -0.6006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9484 -0.6006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4160 -0.8474 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.4160 -0.8474 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.5859 -0.6471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5859 -0.6471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9742 -1.6501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9742 -1.6501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7831 -1.4951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7831 -1.4951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2570 2.1807 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 0.2570 2.1807 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 0.3335 1.5708 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.3335 1.5708 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.9003 1.4835 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.9003 1.4835 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.3497 1.2305 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.3497 1.2305 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.2126 1.7425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2126 1.7425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6842 1.7626 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.6842 1.7626 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.2054 2.7717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2054 2.7717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2006 1.3464 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2006 1.3464 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9085 0.8852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9085 0.8852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6842 1.7626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6842 1.7626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1842 2.6286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1842 2.6286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6410 -2.7373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6410 -2.7373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6410 -3.5623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6410 -3.5623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1085 -0.3050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1085 -0.3050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3907 0.4702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3907 0.4702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 13 22 1 0 0 0 0 | + | 13 22 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 24 20 1 0 0 0 0 | + | 24 20 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 17 1 0 0 0 0 | + | 35 17 1 0 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 44 43 1 1 0 0 0 | + | 44 43 1 1 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 21 1 0 0 0 0 | + | 44 21 1 0 0 0 0 |
| − | 46 50 1 0 0 0 0 | + | 46 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 26 52 1 0 0 0 0 | + | 26 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 37 54 1 0 0 0 0 | + | 37 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 54 55 | + | M SAL 3 2 54 55 |
| − | M SBL 3 1 59 | + | M SBL 3 1 59 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 59 -3.8144 -0.0632 | + | M SVB 3 59 -3.8144 -0.0632 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 57 | + | M SBL 2 1 57 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 57 3.7944 -2.4182 | + | M SVB 2 57 3.7944 -2.4182 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 55 | + | M SBL 1 1 55 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 55 0.5231 2.2473 | + | M SVB 1 55 0.5231 2.2473 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAGGL0014 | + | ID FL7AAGGL0014 |
| − | KNApSAcK_ID C00006719 | + | KNApSAcK_ID C00006719 |
| − | NAME Delphinidin 3,7,3'-triglucoside | + | NAME Delphinidin 3,7,3'-triglucoside |
| − | CAS_RN 64963-58-2 | + | CAS_RN 64963-58-2 |
| − | FORMULA C33H41O22 | + | FORMULA C33H41O22 |
| − | EXACTMASS 789.208947996 | + | EXACTMASS 789.208947996 |
| − | AVERAGEMASS 789.66544 | + | AVERAGEMASS 789.66544 |
| − | SMILES C(O1)(CO)[C@@H]([C@@H]([C@@H]([C@@H]1Oc(c2)cc([o+1]4)c(cc(c4c(c5)cc(O)c(c5O[C@@H]([C@@H](O)6)OC(CO)[C@@H]([C@@H]6O)O)O)O[C@@H](C3O)O[C@H](CO)[C@@H](C(O)3)O)c(O)2)O)O)O | + | SMILES C(O1)(CO)[C@@H]([C@@H]([C@@H]([C@@H]1Oc(c2)cc([o+1]4)c(cc(c4c(c5)cc(O)c(c5O[C@@H]([C@@H](O)6)OC(CO)[C@@H]([C@@H]6O)O)O)O[C@@H](C3O)O[C@H](CO)[C@@H](C(O)3)O)c(O)2)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-1.2169 -0.8609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2169 -1.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6606 -1.8244 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1043 -1.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1043 -0.8609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6606 -0.5397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4520 -1.8244 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0083 -1.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0083 -0.8609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4520 -0.5397 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.5644 -0.5398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1313 -0.8672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6983 -0.5398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6983 0.1149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1313 0.4422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5644 0.1149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7730 -0.5398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2651 0.4421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6606 -2.4666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4244 -2.2240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1313 1.0967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2651 -0.8671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5818 -1.9239 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.2810 -2.4449 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.8595 -2.2796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4414 -2.4449 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.7423 -1.9239 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.1638 -2.0892 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.0231 -2.0736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2053 -1.6146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1764 -2.1745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9950 -0.6989 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.6238 -1.1888 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.0894 -0.9810 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.5736 -0.9754 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.9484 -0.6006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4160 -0.8474 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.5859 -0.6471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9742 -1.6501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7831 -1.4951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2570 2.1807 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
0.3335 1.5708 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.9003 1.4835 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.3497 1.2305 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.2126 1.7425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6842 1.7626 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.2054 2.7717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2006 1.3464 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9085 0.8852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6842 1.7626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1842 2.6286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6410 -2.7373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6410 -3.5623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1085 -0.3050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3907 0.4702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
15 21 1 0 0 0 0
13 22 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
28 30 1 0 0 0 0
27 31 1 0 0 0 0
24 20 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 17 1 0 0 0 0
41 42 1 1 0 0 0
42 43 1 1 0 0 0
44 43 1 1 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
46 41 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 21 1 0 0 0 0
46 50 1 0 0 0 0
50 51 1 0 0 0 0
26 52 1 0 0 0 0
52 53 1 0 0 0 0
37 54 1 0 0 0 0
54 55 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 54 55
M SBL 3 1 59
M SMT 3 CH2OH
M SVB 3 59 -3.8144 -0.0632
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 57
M SMT 2 CH2OH
M SVB 2 57 3.7944 -2.4182
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 55
M SMT 1 CH2OH
M SVB 1 55 0.5231 2.2473
S SKP 8
ID FL7AAGGL0014
KNApSAcK_ID C00006719
NAME Delphinidin 3,7,3'-triglucoside
CAS_RN 64963-58-2
FORMULA C33H41O22
EXACTMASS 789.208947996
AVERAGEMASS 789.66544
SMILES C(O1)(CO)[C@@H]([C@@H]([C@@H]([C@@H]1Oc(c2)cc([o+1]4)c(cc(c4c(c5)cc(O)c(c5O[C@@H]([C@@H](O)6)OC(CO)[C@@H]([C@@H]6O)O)O)O[C@@H](C3O)O[C@H](CO)[C@@H](C(O)3)O)c(O)2)O)O)O
M END