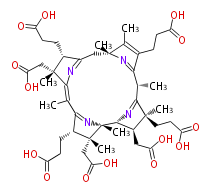
Mol:BMCCPPPC0008
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 63 67 0 0 1 0 0 0 0 0999 V2000 | + | 63 67 0 0 1 0 0 0 0 0999 V2000 |
| − | 6.8290 2.3666 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 6.8290 2.3666 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 5.8509 3.0773 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 5.8509 3.0773 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 4.8727 2.3666 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.8727 2.3666 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 5.2464 1.2167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2464 1.2167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9373 0.2657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9373 0.2657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0419 -0.7288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0419 -0.7288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4373 -1.7759 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 4.4373 -1.7759 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 5.2464 -2.6744 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 5.2464 -2.6744 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 6.3509 -2.1827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.3509 -2.1827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.3290 -2.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.3290 -2.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.3072 -2.1827 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 8.3072 -2.1827 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 9.4117 -2.6744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.4117 -2.6744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.2207 -1.7759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.2207 -1.7759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.6162 -0.7288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.6162 -0.7288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.7207 0.2657 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 9.7207 0.2657 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 9.4117 1.2167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.4117 1.2167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.7853 2.3666 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 9.7853 2.3666 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 8.8072 3.0773 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 8.8072 3.0773 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 7.8290 2.3666 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 7.8290 2.3666 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 5.9155 1.9599 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9155 1.9599 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5419 -1.5949 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5419 -1.5949 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.1162 -1.5949 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.1162 -1.5949 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.7426 1.9599 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.7426 1.9599 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.6381 2.9544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.6381 2.9544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.4387 3.8863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.4387 3.8863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2631 3.8863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2631 3.8863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6698 4.7998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6698 4.7998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9217 2.6756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9217 2.6756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7138 3.6538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7138 3.6538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7627 3.9628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7627 3.9628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9428 0.3702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9428 0.3702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5713 -1.2759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5713 -1.2759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6942 -2.4450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6942 -2.4450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7431 -2.1360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7431 -2.1360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0385 -3.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0385 -3.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0874 -3.9616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0874 -3.9616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8795 -4.9397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8795 -4.9397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.6381 -2.9258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.6381 -2.9258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.6196 -3.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.6196 -3.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.2153 -1.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.2153 -1.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.6220 -2.7940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.6220 -2.7940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.6165 -2.8985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.6165 -2.8985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.7153 0.3702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.7153 0.3702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.7364 2.0576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.7364 2.0576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.3731 3.1756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.3731 3.1756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.3676 3.0711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.3676 3.0711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.9554 3.8801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.9554 3.8801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.8072 4.0773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.8072 4.0773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.6732 4.5773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.6732 4.5773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.2043 -2.0895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.2043 -2.0895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.0232 -3.8121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.0232 -3.8121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6226 -5.6089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6226 -5.6089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9284 -5.2487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9284 -5.2487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5352 -1.1579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5352 -1.1579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0000 -2.8052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0000 -2.8052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0196 3.2937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0196 3.2937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5548 4.9409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5548 4.9409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.5487 4.7937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.5487 4.7937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.9499 3.7756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.9499 3.7756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.5392 4.0773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.5392 4.0773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.6732 5.5773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.6732 5.5773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.6644 4.9044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.6644 4.9044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0821 5.6089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0821 5.6089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 37 52 1 0 0 0 0 | + | 37 52 1 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 37 53 2 0 0 0 0 | + | 37 53 2 0 0 0 0 |
| − | 11 22 1 6 0 0 0 | + | 11 22 1 6 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 34 54 1 0 0 0 0 | + | 34 54 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 34 55 2 0 0 0 0 | + | 34 55 2 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 23 16 2 0 0 0 0 | + | 23 16 2 0 0 0 0 |
| − | 30 56 1 0 0 0 0 | + | 30 56 1 0 0 0 0 |
| − | 17 16 1 0 0 0 0 | + | 17 16 1 0 0 0 0 |
| − | 30 57 2 0 0 0 0 | + | 30 57 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 19 23 1 1 0 0 0 | + | 19 23 1 1 0 0 0 |
| − | 16 15 1 0 0 0 0 | + | 16 15 1 0 0 0 0 |
| − | 15 14 1 0 0 0 0 | + | 15 14 1 0 0 0 0 |
| − | 12 39 1 0 0 0 0 | + | 12 39 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 11 10 1 0 0 0 0 | + | 11 10 1 0 0 0 0 |
| − | 10 9 1 0 0 0 0 | + | 10 9 1 0 0 0 0 |
| − | 6 21 1 0 0 0 0 | + | 6 21 1 0 0 0 0 |
| − | 7 32 1 1 0 0 0 | + | 7 32 1 1 0 0 0 |
| − | 3 28 1 1 0 0 0 | + | 3 28 1 1 0 0 0 |
| − | 47 58 1 0 0 0 0 | + | 47 58 1 0 0 0 0 |
| − | 9 8 1 0 0 0 0 | + | 9 8 1 0 0 0 0 |
| − | 47 59 2 0 0 0 0 | + | 47 59 2 0 0 0 0 |
| − | 8 7 1 0 0 0 0 | + | 8 7 1 0 0 0 0 |
| − | 18 48 1 6 0 0 0 | + | 18 48 1 6 0 0 0 |
| − | 7 6 1 0 0 0 0 | + | 7 6 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 21 9 2 0 0 0 0 | + | 21 9 2 0 0 0 0 |
| − | 49 60 1 0 0 0 0 | + | 49 60 1 0 0 0 0 |
| − | 2 1 1 0 0 0 0 | + | 2 1 1 0 0 0 0 |
| − | 49 61 2 0 0 0 0 | + | 49 61 2 0 0 0 0 |
| − | 13 40 1 0 0 0 0 | + | 13 40 1 0 0 0 0 |
| − | 20 4 2 0 0 0 0 | + | 20 4 2 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 3 2 1 0 0 0 0 | + | 3 2 1 0 0 0 0 |
| − | 1 20 1 6 0 0 0 | + | 1 20 1 6 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 17 44 1 6 0 0 0 | + | 17 44 1 6 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 7 33 1 6 0 0 0 | + | 7 33 1 6 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 8 35 1 1 0 0 0 | + | 8 35 1 1 0 0 0 |
| − | 2 26 1 6 0 0 0 | + | 2 26 1 6 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 2 25 1 1 0 0 0 | + | 2 25 1 1 0 0 0 |
| − | 42 50 2 0 0 0 0 | + | 42 50 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 14 22 2 0 0 0 0 | + | 14 22 2 0 0 0 0 |
| − | 27 62 1 0 0 0 0 | + | 27 62 1 0 0 0 0 |
| − | 42 51 1 0 0 0 0 | + | 42 51 1 0 0 0 0 |
| − | 27 63 2 0 0 0 0 | + | 27 63 2 0 0 0 0 |
| − | 11 38 1 1 0 0 0 | + | 11 38 1 1 0 0 0 |
| − | 1 24 1 1 0 0 0 | + | 1 24 1 1 0 0 0 |
| − | 17 45 1 1 0 0 0 | + | 17 45 1 1 0 0 0 |
| − | 5 31 1 0 0 0 0 | + | 5 31 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 15 43 1 4 0 0 0 | + | 15 43 1 4 0 0 0 |
| − | S SKP 7 | + | S SKP 7 |
| − | ID BMCCPPPC0008 | + | ID BMCCPPPC0008 |
| − | NAME Precorrin 8X | + | NAME Precorrin 8X |
| − | FORMULA C45H60N4O14 | + | FORMULA C45H60N4O14 |
| − | EXACTMASS 880.4106 | + | EXACTMASS 880.4106 |
| − | AVERAGEMASS 880.9764 | + | AVERAGEMASS 880.9764 |
| − | SMILES C(CCC(C3=5)=C(C)[C@@](N5)(C)CC([C@H]4CCC(O)=O)=NC([C@]4(C)CC(O)=O)=C(C(=N1)[C@H]([C@](CC(O)=O)(C)[C@@]([C@H](N=2)[C@H](CC(O)=O)[C@](CCC(O)=O)(C2C3C)C)1C)CCC(O)=O)C)(O)=O | + | SMILES C(CCC(C3=5)=C(C)[C@@](N5)(C)CC([C@H]4CCC(O)=O)=NC([C@]4(C)CC(O)=O)=C(C(=N1)[C@H]([C@](CC(O)=O)(C)[C@@]([C@H](N=2)[C@H](CC(O)=O)[C@](CCC(O)=O)(C2C3C)C)1C)CCC(O)=O)C)(O)=O |
| − | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C06408 | + | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C06408 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
63 67 0 0 1 0 0 0 0 0999 V2000
6.8290 2.3666 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
5.8509 3.0773 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
4.8727 2.3666 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
5.2464 1.2167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9373 0.2657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0419 -0.7288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4373 -1.7759 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
5.2464 -2.6744 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
6.3509 -2.1827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.3290 -2.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.3072 -2.1827 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
9.4117 -2.6744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.2207 -1.7759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.6162 -0.7288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.7207 0.2657 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
9.4117 1.2167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.7853 2.3666 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
8.8072 3.0773 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
7.8290 2.3666 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
5.9155 1.9599 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
5.5419 -1.5949 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
9.1162 -1.5949 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
8.7426 1.9599 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
7.6381 2.9544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.4387 3.8863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2631 3.8863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6698 4.7998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9217 2.6756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7138 3.6538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7627 3.9628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9428 0.3702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5713 -1.2759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6942 -2.4450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7431 -2.1360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0385 -3.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0874 -3.9616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8795 -4.9397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.6381 -2.9258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.6196 -3.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.2153 -1.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.6220 -2.7940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
12.6165 -2.8985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.7153 0.3702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.7364 2.0576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.3731 3.1756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.3676 3.0711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.9554 3.8801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.8072 4.0773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.6732 4.5773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
13.2043 -2.0895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
13.0232 -3.8121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6226 -5.6089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9284 -5.2487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5352 -1.1579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0000 -2.8052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0196 3.2937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5548 4.9409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
11.5487 4.7937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
12.9499 3.7756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
10.5392 4.0773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.6732 5.5773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.6644 4.9044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0821 5.6089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
36 37 1 0 0 0 0
12 13 2 0 0 0 0
37 52 1 0 0 0 0
13 14 1 0 0 0 0
37 53 2 0 0 0 0
11 22 1 6 0 0 0
33 34 1 0 0 0 0
18 19 1 0 0 0 0
34 54 1 0 0 0 0
28 29 1 0 0 0 0
34 55 2 0 0 0 0
45 46 1 0 0 0 0
29 30 1 0 0 0 0
23 16 2 0 0 0 0
30 56 1 0 0 0 0
17 16 1 0 0 0 0
30 57 2 0 0 0 0
17 18 1 0 0 0 0
19 23 1 1 0 0 0
16 15 1 0 0 0 0
15 14 1 0 0 0 0
12 39 1 0 0 0 0
46 47 1 0 0 0 0
11 10 1 0 0 0 0
10 9 1 0 0 0 0
6 21 1 0 0 0 0
7 32 1 1 0 0 0
3 28 1 1 0 0 0
47 58 1 0 0 0 0
9 8 1 0 0 0 0
47 59 2 0 0 0 0
8 7 1 0 0 0 0
18 48 1 6 0 0 0
7 6 1 0 0 0 0
48 49 1 0 0 0 0
21 9 2 0 0 0 0
49 60 1 0 0 0 0
2 1 1 0 0 0 0
49 61 2 0 0 0 0
13 40 1 0 0 0 0
20 4 2 0 0 0 0
4 3 1 0 0 0 0
3 2 1 0 0 0 0
1 20 1 6 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
17 44 1 6 0 0 0
1 19 1 0 0 0 0
40 41 1 0 0 0 0
7 33 1 6 0 0 0
41 42 1 0 0 0 0
8 35 1 1 0 0 0
2 26 1 6 0 0 0
35 36 1 0 0 0 0
2 25 1 1 0 0 0
42 50 2 0 0 0 0
26 27 1 0 0 0 0
14 22 2 0 0 0 0
27 62 1 0 0 0 0
42 51 1 0 0 0 0
27 63 2 0 0 0 0
11 38 1 1 0 0 0
1 24 1 1 0 0 0
17 45 1 1 0 0 0
5 31 1 0 0 0 0
11 12 1 0 0 0 0
15 43 1 4 0 0 0
S SKP 7
ID BMCCPPPC0008
NAME Precorrin 8X
FORMULA C45H60N4O14
EXACTMASS 880.4106
AVERAGEMASS 880.9764
SMILES C(CCC(C3=5)=C(C)[C@@](N5)(C)CC([C@H]4CCC(O)=O)=NC([C@]4(C)CC(O)=O)=C(C(=N1)[C@H]([C@](CC(O)=O)(C)[C@@]([C@H](N=2)[C@H](CC(O)=O)[C@](CCC(O)=O)(C2C3C)C)1C)CCC(O)=O)C)(O)=O
KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C06408
M END