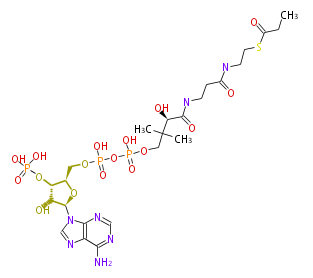
Mol:BMFYS3ESa001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 52 54 0 0 1 0 0 0 0 0999 V2000 | + | 52 54 0 0 1 0 0 0 0 0999 V2000 |
| − | 3.6473 -3.8348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6473 -3.8348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6691 -3.6269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6691 -3.6269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0000 -4.3701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0000 -4.3701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9563 -4.7859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9563 -4.7859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21.0044 -2.8049 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 21.0044 -2.8049 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 20.1384 -2.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 20.1384 -2.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 20.1384 -1.3049 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 20.1384 -1.3049 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21.0044 -0.8049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 21.0044 -0.8049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21.8704 -1.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 21.8704 -1.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21.8704 -2.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 21.8704 -2.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.6136 -0.6358 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.6136 -0.6358 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.2068 0.2778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.2068 0.2778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21.2123 0.1733 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 21.2123 0.1733 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.7364 -2.8049 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.7364 -2.8049 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 20.5432 0.9164 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 20.5432 0.9164 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 20.7511 1.8946 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 20.7511 1.8946 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 19.8851 2.3946 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 19.8851 2.3946 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 19.1419 1.7254 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 19.1419 1.7254 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 18.1638 1.9333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.1638 1.9333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21.6646 2.3013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 21.6646 2.3013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.7805 3.3891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.7805 3.3891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.5486 0.8119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.5486 0.8119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 17.4946 1.1902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 17.4946 1.1902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 20.5895 3.9769 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 20.5895 3.9769 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21.1773 3.1678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 21.1773 3.1678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 20.0018 4.7859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 20.0018 4.7859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21.3986 4.5646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 21.3986 4.5646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.5165 1.3981 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 16.5165 1.3981 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.3086 0.4200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 16.3086 0.4200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.7244 2.3762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 16.7244 2.3762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 15.5383 1.6060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 15.5383 1.6060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.8692 0.8629 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.8692 0.8629 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 15.6124 0.1937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 15.6124 0.1937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.1261 1.5320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.1261 1.5320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.2001 0.1197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.2001 0.1197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.2219 0.3276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.2219 0.3276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.5528 -0.4155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.5528 -0.4155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.2959 -1.0846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.2959 -1.0846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.8097 0.2536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.8097 0.2536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.8837 -1.1587 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 11.8837 -1.1587 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 10.9055 -0.9507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.9055 -0.9507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.2364 -1.6939 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.2364 -1.6939 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.2582 -1.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.2582 -1.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.5891 -2.2291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.5891 -2.2291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.6110 -2.0212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.6110 -2.0212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.9418 -2.7644 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.9418 -2.7644 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9637 -2.5564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9637 -2.5564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2946 -3.2996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2946 -3.2996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3164 -3.0917 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3164 -3.0917 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.1927 -2.1097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.1927 -2.1097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.5965 0.0003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.5965 0.0003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.3019 -1.0702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.3019 -1.0702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5 10 2 0 0 0 0 | + | 5 10 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 2 0 0 0 0 | + | 6 7 2 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 9 8 2 0 0 0 0 | + | 9 8 2 0 0 0 0 |
| − | 13 12 1 0 0 0 0 | + | 13 12 1 0 0 0 0 |
| − | 12 11 2 0 0 0 0 | + | 12 11 2 0 0 0 0 |
| − | 11 9 1 0 0 0 0 | + | 11 9 1 0 0 0 0 |
| − | 8 13 1 0 0 0 0 | + | 8 13 1 0 0 0 0 |
| − | 10 14 1 0 0 0 0 | + | 10 14 1 0 0 0 0 |
| − | 18 22 1 6 0 0 0 | + | 18 22 1 6 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 15 22 1 6 0 0 0 | + | 15 22 1 6 0 0 0 |
| − | 17 16 1 0 0 0 0 | + | 17 16 1 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 19 23 1 0 0 0 0 | + | 19 23 1 0 0 0 0 |
| − | 15 13 1 0 0 0 0 | + | 15 13 1 0 0 0 0 |
| − | 23 28 1 0 0 0 0 | + | 23 28 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 28 29 2 0 0 0 0 | + | 28 29 2 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 35 32 1 0 0 0 0 | + | 35 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 32 31 1 0 0 0 0 | + | 32 31 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 40 50 1 1 0 0 0 | + | 40 50 1 1 0 0 0 |
| − | 41 51 2 0 0 0 0 | + | 41 51 2 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 45 52 2 0 0 0 0 | + | 45 52 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 16 20 1 1 0 0 0 | + | 16 20 1 1 0 0 0 |
| − | 17 21 1 1 0 0 0 | + | 17 21 1 1 0 0 0 |
| − | 25 24 1 0 0 0 0 | + | 25 24 1 0 0 0 0 |
| − | 24 27 1 0 0 0 0 | + | 24 27 1 0 0 0 0 |
| − | 24 26 2 0 0 0 0 | + | 24 26 2 0 0 0 0 |
| − | 21 24 1 0 0 0 0 | + | 21 24 1 0 0 0 0 |
| − | 49 1 1 0 0 0 0 | + | 49 1 1 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 1 4 2 0 0 0 0 | + | 1 4 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | S SKP 7 | + | S SKP 7 |
| − | ID BMFYS3ESa001 | + | ID BMFYS3ESa001 |
| − | NAME Propanoyl-CoA | + | NAME Propanoyl-CoA |
| − | FORMULA C24H40N7O17P3S | + | FORMULA C24H40N7O17P3S |
| − | EXACTMASS 823.1414 | + | EXACTMASS 823.1414 |
| − | AVERAGEMASS 823.5986 | + | AVERAGEMASS 823.5986 |
| − | SMILES n(c3N)cnc(c32)n(cn2)[C@@H]([C@@H]1O)O[C@@H]([C@H]1OP(O)(O)=O)COP(OP(O)(=O)OCC([C@@H](O)C(NCCC(=O)NCCSC(CC)=O)=O)(C)C)(O)=O | + | SMILES n(c3N)cnc(c32)n(cn2)[C@@H]([C@@H]1O)O[C@@H]([C@H]1OP(O)(O)=O)COP(OP(O)(=O)OCC([C@@H](O)C(NCCC(=O)NCCSC(CC)=O)=O)(C)C)(O)=O |
| − | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C00100 | + | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C00100 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
52 54 0 0 1 0 0 0 0 0999 V2000
3.6473 -3.8348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6691 -3.6269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0000 -4.3701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9563 -4.7859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
21.0044 -2.8049 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
20.1384 -2.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
20.1384 -1.3049 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
21.0044 -0.8049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
21.8704 -1.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
21.8704 -2.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
22.6136 -0.6358 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
22.2068 0.2778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
21.2123 0.1733 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
22.7364 -2.8049 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
20.5432 0.9164 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
20.7511 1.8946 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
19.8851 2.3946 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
19.1419 1.7254 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
18.1638 1.9333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
21.6646 2.3013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
19.7805 3.3891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
19.5486 0.8119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
17.4946 1.1902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
20.5895 3.9769 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
21.1773 3.1678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
20.0018 4.7859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
21.3986 4.5646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
16.5165 1.3981 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
16.3086 0.4200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
16.7244 2.3762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
15.5383 1.6060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
14.8692 0.8629 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
15.6124 0.1937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
14.1261 1.5320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
14.2001 0.1197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
13.2219 0.3276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
12.5528 -0.4155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
13.2959 -1.0846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.8097 0.2536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.8837 -1.1587 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
10.9055 -0.9507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.2364 -1.6939 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
9.2582 -1.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.5891 -2.2291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.6110 -2.0212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.9418 -2.7644 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
5.9637 -2.5564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2946 -3.2996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3164 -3.0917 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0
12.1927 -2.1097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
10.5965 0.0003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.3019 -1.0702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5 10 2 0 0 0 0
5 6 1 0 0 0 0
6 7 2 0 0 0 0
7 8 1 0 0 0 0
9 10 1 0 0 0 0
9 8 2 0 0 0 0
13 12 1 0 0 0 0
12 11 2 0 0 0 0
11 9 1 0 0 0 0
8 13 1 0 0 0 0
10 14 1 0 0 0 0
18 22 1 6 0 0 0
17 18 1 0 0 0 0
15 22 1 6 0 0 0
17 16 1 0 0 0 0
15 16 1 0 0 0 0
18 19 1 0 0 0 0
19 23 1 0 0 0 0
15 13 1 0 0 0 0
23 28 1 0 0 0 0
28 30 1 0 0 0 0
28 29 2 0 0 0 0
28 31 1 0 0 0 0
35 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
35 36 1 0 0 0 0
32 31 1 0 0 0 0
36 37 1 0 0 0 0
37 40 1 0 0 0 0
37 38 1 0 0 0 0
37 39 1 0 0 0 0
40 41 1 0 0 0 0
40 50 1 1 0 0 0
41 51 2 0 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
45 52 2 0 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 49 1 0 0 0 0
16 20 1 1 0 0 0
17 21 1 1 0 0 0
25 24 1 0 0 0 0
24 27 1 0 0 0 0
24 26 2 0 0 0 0
21 24 1 0 0 0 0
49 1 1 0 0 0 0
1 2 1 0 0 0 0
1 4 2 0 0 0 0
2 3 1 0 0 0 0
S SKP 7
ID BMFYS3ESa001
NAME Propanoyl-CoA
FORMULA C24H40N7O17P3S
EXACTMASS 823.1414
AVERAGEMASS 823.5986
SMILES n(c3N)cnc(c32)n(cn2)[C@@H]([C@@H]1O)O[C@@H]([C@H]1OP(O)(O)=O)COP(OP(O)(=O)OCC([C@@H](O)C(NCCC(=O)NCCSC(CC)=O)=O)(C)C)(O)=O
KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C00100
M END