Mol:BMMCHC--o020
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 56 0 0 1 0 0 0 0 0999 V2000 | + | 55 56 0 0 1 0 0 0 0 0999 V2000 |
| − | -2.1274 2.2150 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.1274 2.2150 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -1.4129 2.6275 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -1.4129 2.6275 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -2.1290 0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1290 0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8419 2.6275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8419 2.6275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8391 3.4523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8391 3.4523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5550 2.2125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5550 2.2125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4129 3.4524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4129 3.4524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6973 3.8625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6973 3.8625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1289 3.8626 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1289 3.8626 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6985 2.2150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6985 2.2150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0159 2.6275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0159 2.6275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4149 -0.1182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4149 -0.1182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7001 0.2937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7001 0.2937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0140 -0.1194 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0140 -0.1194 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6995 1.1187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6995 1.1187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3558 0.0654 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.3558 0.0654 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 2.1808 0.0654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1808 0.0654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3551 0.8904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3551 0.8904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0704 1.3011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0704 1.3011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6388 1.2999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6388 1.2999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9031 -0.3333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9031 -0.3333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6095 0.0927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6095 0.0927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4692 -0.5811 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4692 -0.5811 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6121 0.8718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6121 0.8718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7237 -0.3517 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 5.7237 -0.3517 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 5.7235 0.5100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7235 0.5100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.4391 0.9202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.4391 0.9202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0350 1.0667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0350 1.0667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7545 -1.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7545 -1.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4712 -1.7715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4712 -1.7715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7574 -1.3579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7574 -1.3579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0423 -1.7694 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0423 -1.7694 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3284 -1.3559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3284 -1.3559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7585 -0.5329 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7585 -0.5329 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3436 -0.6346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3436 -0.6346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7614 -1.3565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7614 -1.3565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3542 -2.0793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3542 -2.0793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4832 -2.0768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4832 -2.0768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9011 -1.3549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9011 -1.3549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4813 -0.6354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4813 -0.6354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5864 -1.3518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5864 -1.3518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4116 -1.3508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4116 -1.3508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8250 -2.0647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8250 -2.0647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6485 -2.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6485 -2.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8154 -2.9587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8154 -2.9587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1090 -3.3695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1090 -3.3695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4896 -2.8220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4896 -2.8220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5686 -3.2951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5686 -3.2951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6859 -2.7743 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6859 -2.7743 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6134 -1.7674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6134 -1.7674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9091 2.2882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9091 2.2882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1081 1.5917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1081 1.5917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6224 2.7031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6224 2.7031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.4391 -3.1108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.4391 -3.1108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.4235 -3.8626 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.4235 -3.8626 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 1 3 1 4 0 0 0 | + | 1 3 1 4 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 4 6 2 0 0 0 0 | + | 4 6 2 0 0 0 0 |
| − | 1 4 1 0 0 0 0 | + | 1 4 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 7 9 2 0 0 0 0 | + | 7 9 2 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 2 10 1 4 0 0 0 | + | 2 10 1 4 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 13 15 2 0 0 0 0 | + | 13 15 2 0 0 0 0 |
| − | 16 14 1 1 0 0 0 | + | 16 14 1 1 0 0 0 |
| − | 16 17 1 1 0 0 0 | + | 16 17 1 1 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 18 20 2 0 0 0 0 | + | 18 20 2 0 0 0 0 |
| − | 16 18 1 0 0 0 0 | + | 16 18 1 0 0 0 0 |
| − | 17 21 1 0 0 0 0 | + | 17 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 22 24 2 0 0 0 0 | + | 22 24 2 0 0 0 0 |
| − | 25 23 1 1 0 0 0 | + | 25 23 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 26 28 2 0 0 0 0 | + | 26 28 2 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 31 34 2 0 0 0 0 | + | 31 34 2 0 0 0 0 |
| − | 35 40 2 0 0 0 0 | + | 35 40 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 36 41 1 0 0 0 0 | + | 36 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 43 2 0 0 0 0 | + | 47 43 2 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 45 48 1 0 0 0 0 | + | 45 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 33 50 1 0 0 0 0 | + | 33 50 1 0 0 0 0 |
| − | 50 39 1 0 0 0 0 | + | 50 39 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 51 53 2 0 0 0 0 | + | 51 53 2 0 0 0 0 |
| − | 11 51 1 0 0 0 0 | + | 11 51 1 0 0 0 0 |
| − | 49 54 1 0 0 0 0 | + | 49 54 1 0 0 0 0 |
| − | 54 55 2 0 0 0 0 | + | 54 55 2 0 0 0 0 |
| − | S SKP 7 | + | S SKP 7 |
| − | ID BMMCHC--o020 | + | ID BMMCHC--o020 |
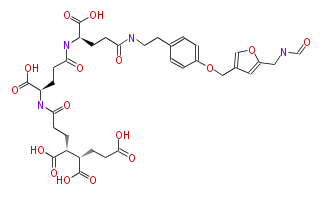
| − | NAME Formyl-methanofuran | + | NAME Formyl-methanofuran |
| − | FORMULA C35H44N4O16 | + | FORMULA C35H44N4O16 |
| − | EXACTMASS 776.2752 | + | EXACTMASS 776.2752 |
| − | AVERAGEMASS 776.7412 | + | AVERAGEMASS 776.7412 |
| − | SMILES N(C(CC[C@@H](NC(=O)CC[C@@H](NC(=O)CCC(C(CCC(O)=O)C(O)=O)C(O)=O)C(O)=O)C(O)=O)=O)CCc(c1)ccc(OCc(c2)cc(CNC=O)o2)c1 | + | SMILES N(C(CC[C@@H](NC(=O)CC[C@@H](NC(=O)CCC(C(CCC(O)=O)C(O)=O)C(O)=O)C(O)=O)C(O)=O)=O)CCc(c1)ccc(OCc(c2)cc(CNC=O)o2)c1 |
| − | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C01001 | + | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C01001 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 56 0 0 1 0 0 0 0 0999 V2000
-2.1274 2.2150 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-1.4129 2.6275 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-2.1290 0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8419 2.6275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8391 3.4523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5550 2.2125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4129 3.4524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6973 3.8625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1289 3.8626 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6985 2.2150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0159 2.6275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4149 -0.1182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7001 0.2937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0140 -0.1194 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-0.6995 1.1187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3558 0.0654 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.1808 0.0654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3551 0.8904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0704 1.3011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6388 1.2999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9031 -0.3333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6095 0.0927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4692 -0.5811 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.6121 0.8718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7237 -0.3517 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
5.7235 0.5100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.4391 0.9202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0350 1.0667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7545 -1.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4712 -1.7715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7574 -1.3579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0423 -1.7694 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
2.3284 -1.3559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7585 -0.5329 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3436 -0.6346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7614 -1.3565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3542 -2.0793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4832 -2.0768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9011 -1.3549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4813 -0.6354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5864 -1.3518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4116 -1.3508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8250 -2.0647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6485 -2.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8154 -2.9587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1090 -3.3695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4896 -2.8220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5686 -3.2951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6859 -2.7743 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
1.6134 -1.7674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9091 2.2882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1081 1.5917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6224 2.7031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.4391 -3.1108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.4235 -3.8626 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
1 3 1 4 0 0 0
4 5 1 0 0 0 0
4 6 2 0 0 0 0
1 4 1 0 0 0 0
7 8 1 0 0 0 0
7 9 2 0 0 0 0
2 7 1 0 0 0 0
2 10 1 4 0 0 0
10 11 1 0 0 0 0
3 12 1 0 0 0 0
12 13 1 0 0 0 0
13 14 1 0 0 0 0
13 15 2 0 0 0 0
16 14 1 1 0 0 0
16 17 1 1 0 0 0
18 19 1 0 0 0 0
18 20 2 0 0 0 0
16 18 1 0 0 0 0
17 21 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 0 0 0 0
22 24 2 0 0 0 0
25 23 1 1 0 0 0
26 27 1 0 0 0 0
26 28 2 0 0 0 0
25 26 1 0 0 0 0
25 29 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
31 34 2 0 0 0 0
35 40 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
36 41 1 0 0 0 0
41 42 1 0 0 0 0
43 44 1 0 0 0 0
44 45 2 0 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 43 2 0 0 0 0
42 43 1 0 0 0 0
45 48 1 0 0 0 0
48 49 1 0 0 0 0
33 50 1 0 0 0 0
50 39 1 0 0 0 0
51 52 1 0 0 0 0
51 53 2 0 0 0 0
11 51 1 0 0 0 0
49 54 1 0 0 0 0
54 55 2 0 0 0 0
S SKP 7
ID BMMCHC--o020
NAME Formyl-methanofuran
FORMULA C35H44N4O16
EXACTMASS 776.2752
AVERAGEMASS 776.7412
SMILES N(C(CC[C@@H](NC(=O)CC[C@@H](NC(=O)CCC(C(CCC(O)=O)C(O)=O)C(O)=O)C(O)=O)C(O)=O)=O)CCc(c1)ccc(OCc(c2)cc(CNC=O)o2)c1
KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C01001
M END