Mol:FL1CALNI0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 33 34 0 0 0 0 0 0 0 0999 V2000 | + | 33 34 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.9455 0.1449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9455 0.1449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9455 -0.4975 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9455 -0.4975 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5018 -0.8187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5018 -0.8187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0581 -0.4975 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0581 -0.4975 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0581 0.1449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0581 0.1449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5018 0.4661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5018 0.4661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8123 -0.4769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8123 -0.4769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3686 -0.7980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3686 -0.7980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2562 -0.7979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2562 -0.7979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3048 -0.4643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3048 -0.4643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2562 -1.4387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2562 -1.4387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3048 0.1738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3048 0.1738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8574 0.4929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8574 0.4929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4100 0.1738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4100 0.1738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4100 -0.4643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4100 -0.4643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8574 -0.7834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8574 -0.7834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8574 -1.4214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8574 -1.4214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9626 0.4928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9626 0.4928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6142 0.4659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6142 0.4659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9626 -0.7833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9626 -0.7833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5139 -0.4650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5139 -0.4650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5139 0.1702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5139 0.1702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0640 -0.7826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0640 -0.7826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0640 -1.4179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0640 -1.4179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6142 -0.4650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6142 -0.4650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0640 0.4879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0640 0.4879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0640 1.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0640 1.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6130 1.4387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6130 1.4387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5151 1.4387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5151 1.4387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0654 -0.9792 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0654 -0.9792 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7798 -1.3917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7798 -1.3917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2478 0.4928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2478 0.4928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9622 0.0803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9622 0.0803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 7 9 1 0 0 0 0 | + | 7 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 9 11 2 0 0 0 0 | + | 9 11 2 0 0 0 0 |
| − | 2 8 1 0 0 0 0 | + | 2 8 1 0 0 0 0 |
| − | 10 12 2 0 0 0 0 | + | 10 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 5 19 1 0 0 0 0 | + | 5 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 21 23 1 0 0 0 0 | + | 21 23 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 22 26 1 0 0 0 0 | + | 22 26 1 0 0 0 0 |
| − | 26 27 2 0 0 0 0 | + | 26 27 2 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 3 30 1 0 0 0 0 | + | 3 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 12 32 1 0 0 0 0 | + | 12 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 30 31 | + | M SAL 1 2 30 31 |
| − | M SBL 1 1 31 | + | M SBL 1 1 31 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 31 -8.9643 2.0468 | + | M SBV 1 31 -8.9643 2.0468 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 32 33 | + | M SAL 2 2 32 33 |
| − | M SBL 2 1 33 | + | M SBL 2 1 33 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 33 -7.9754 2.5264 | + | M SBV 2 33 -7.9754 2.5264 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL1CALNI0001 | + | ID FL1CALNI0001 |
| − | KNApSAcK_ID C00007139 | + | KNApSAcK_ID C00007139 |
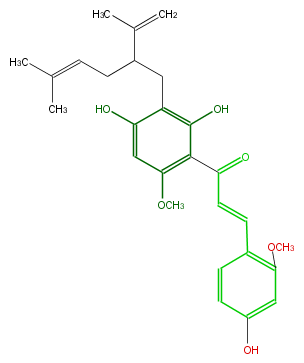
| − | NAME Kushenol D | + | NAME Kushenol D |
| − | CAS_RN 99217-65-9 | + | CAS_RN 99217-65-9 |
| − | FORMULA C27H32O6 | + | FORMULA C27H32O6 |
| − | EXACTMASS 452.219888756 | + | EXACTMASS 452.219888756 |
| − | AVERAGEMASS 452.53938 | + | AVERAGEMASS 452.53938 |
| − | SMILES c(c1)c(C=CC(=O)c(c2O)c(OC)cc(O)c(CC(C(C)=C)CC=C(C)C)2)c(OC)cc1O | + | SMILES c(c1)c(C=CC(=O)c(c2O)c(OC)cc(O)c(CC(C(C)=C)CC=C(C)C)2)c(OC)cc1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
33 34 0 0 0 0 0 0 0 0999 V2000
1.9455 0.1449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9455 -0.4975 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5018 -0.8187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0581 -0.4975 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0581 0.1449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5018 0.4661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8123 -0.4769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3686 -0.7980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2562 -0.7979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3048 -0.4643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2562 -1.4387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3048 0.1738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8574 0.4929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4100 0.1738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4100 -0.4643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8574 -0.7834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8574 -1.4214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9626 0.4928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6142 0.4659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9626 -0.7833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5139 -0.4650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5139 0.1702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0640 -0.7826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0640 -1.4179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6142 -0.4650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0640 0.4879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0640 1.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6130 1.4387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5151 1.4387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0654 -0.9792 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7798 -1.3917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2478 0.4928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9622 0.0803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
7 8 2 0 0 0 0
7 9 1 0 0 0 0
9 10 1 0 0 0 0
9 11 2 0 0 0 0
2 8 1 0 0 0 0
10 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
16 17 1 0 0 0 0
14 18 1 0 0 0 0
5 19 1 0 0 0 0
15 20 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 0 0 0 0
21 23 1 0 0 0 0
23 24 2 0 0 0 0
23 25 1 0 0 0 0
22 26 1 0 0 0 0
26 27 2 0 0 0 0
27 28 1 0 0 0 0
27 29 1 0 0 0 0
3 30 1 0 0 0 0
30 31 1 0 0 0 0
12 32 1 0 0 0 0
32 33 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 30 31
M SBL 1 1 31
M SMT 1 OCH3
M SBV 1 31 -8.9643 2.0468
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 32 33
M SBL 2 1 33
M SMT 2 OCH3
M SBV 2 33 -7.9754 2.5264
S SKP 8
ID FL1CALNI0001
KNApSAcK_ID C00007139
NAME Kushenol D
CAS_RN 99217-65-9
FORMULA C27H32O6
EXACTMASS 452.219888756
AVERAGEMASS 452.53938
SMILES c(c1)c(C=CC(=O)c(c2O)c(OC)cc(O)c(CC(C(C)=C)CC=C(C)C)2)c(OC)cc1O
M END