Mol:FL1CQUCS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.2353 0.4180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2353 0.4180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2353 -0.2908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2353 -0.2908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9779 -0.7196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9779 -0.7196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9779 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9779 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2353 -2.0056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2353 -2.0056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5073 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5073 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5073 -0.7196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5073 -0.7196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2353 -2.6663 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2353 -2.6663 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1605 -0.0623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1605 -0.0623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9973 -0.4365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9973 -0.4365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2658 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2658 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6245 -0.3462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6245 -0.3462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2658 -0.6734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2658 -0.6734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0482 -0.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0482 -0.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8307 -0.6734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8307 -0.6734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8307 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8307 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0482 -2.0286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0482 -2.0286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0482 0.3608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0482 0.3608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3125 -0.3952 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3125 -0.3952 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2146 -2.0332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2146 -2.0332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2146 -2.8582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2146 -2.8582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8932 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8932 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8234 -2.1776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8234 -2.1776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6154 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6154 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2582 -1.9480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2582 -1.9480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9012 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9012 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9012 -0.8345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9012 -0.8345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2582 -0.4633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2582 -0.4633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6154 -0.8345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6154 -0.8345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3941 -0.5500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3941 -0.5500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0714 1.1838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0714 1.1838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5862 1.6819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5862 1.6819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8981 2.4457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8981 2.4457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2050 1.6819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2050 1.6819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5474 1.1838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5474 1.1838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6126 2.8582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6126 2.8582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6175 2.3964 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6175 2.3964 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1307 0.6005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1307 0.6005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7557 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7557 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3941 -0.4711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3941 -0.4711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1737 2.3964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1737 2.3964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1031 2.3964 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1031 2.3964 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 2 0 0 0 0 | + | 6 7 2 0 0 0 0 |
| − | 7 2 1 0 0 0 0 | + | 7 2 1 0 0 0 0 |
| − | 5 8 2 0 0 0 0 | + | 5 8 2 0 0 0 0 |
| − | 2 9 1 0 0 0 0 | + | 2 9 1 0 0 0 0 |
| − | 7 10 1 0 0 0 0 | + | 7 10 1 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 11 13 1 0 0 0 0 | + | 11 13 1 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 11 1 0 0 0 0 | + | 17 11 1 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 6 20 1 0 0 0 0 | + | 6 20 1 0 0 0 0 |
| − | 20 21 2 0 0 0 0 | + | 20 21 2 0 0 0 0 |
| − | 20 22 1 0 0 0 0 | + | 20 22 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 24 2 0 0 0 0 | + | 29 24 2 0 0 0 0 |
| − | 27 30 1 0 0 0 0 | + | 27 30 1 0 0 0 0 |
| − | 1 31 1 0 0 0 0 | + | 1 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 1 35 1 1 0 0 0 | + | 1 35 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 33 36 1 0 0 0 0 | + | 33 36 1 0 0 0 0 |
| − | 34 37 1 0 0 0 0 | + | 34 37 1 0 0 0 0 |
| − | 35 38 1 0 0 0 0 | + | 35 38 1 0 0 0 0 |
| − | 11 4 1 6 0 0 0 | + | 11 4 1 6 0 0 0 |
| − | 14 18 1 6 0 0 0 | + | 14 18 1 6 0 0 0 |
| − | 15 19 1 1 0 0 0 | + | 15 19 1 1 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 16 39 1 0 0 0 0 | + | 16 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 32 41 1 0 0 0 0 | + | 32 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 44 -38.3394 23.9234 | + | M SBV 1 44 -38.3394 23.9234 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 46 -37.0020 24.6378 | + | M SBV 2 46 -37.0020 24.6378 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL1CQUCS0001 | + | ID FL1CQUCS0001 |
| − | KNApSAcK_ID C00006246 | + | KNApSAcK_ID C00006246 |
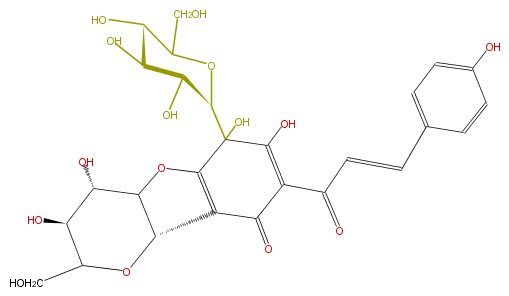
| − | NAME Safflor yellow A | + | NAME Safflor yellow A |
| − | CAS_RN 85532-77-0 | + | CAS_RN 85532-77-0 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES C(CO)(C5O)OC(C(O)C5O)C(C=13)(O)C(O)=C(C(C=Cc(c4)ccc(O)c4)=O)C(C1C(C(O3)2)OC(CO)C(O)C(O)2)=O | + | SMILES C(CO)(C5O)OC(C(O)C5O)C(C=13)(O)C(O)=C(C(C=Cc(c4)ccc(O)c4)=O)C(C1C(C(O3)2)OC(CO)C(O)C(O)2)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.2353 0.4180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2353 -0.2908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9779 -0.7196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9779 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2353 -2.0056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5073 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5073 -0.7196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2353 -2.6663 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1605 -0.0623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9973 -0.4365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2658 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6245 -0.3462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2658 -0.6734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0482 -0.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8307 -0.6734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8307 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0482 -2.0286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0482 0.3608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3125 -0.3952 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2146 -2.0332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2146 -2.8582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8932 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8234 -2.1776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6154 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2582 -1.9480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9012 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9012 -0.8345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2582 -0.4633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6154 -0.8345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3941 -0.5500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0714 1.1838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5862 1.6819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8981 2.4457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2050 1.6819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5474 1.1838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6126 2.8582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6175 2.3964 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1307 0.6005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7557 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3941 -0.4711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1737 2.3964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1031 2.3964 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
6 7 2 0 0 0 0
7 2 1 0 0 0 0
5 8 2 0 0 0 0
2 9 1 0 0 0 0
7 10 1 0 0 0 0
3 12 1 0 0 0 0
11 13 1 0 0 0 0
13 14 1 0 0 0 0
14 15 1 0 0 0 0
15 16 1 0 0 0 0
16 17 1 0 0 0 0
17 11 1 0 0 0 0
12 13 1 0 0 0 0
6 20 1 0 0 0 0
20 21 2 0 0 0 0
20 22 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 29 1 0 0 0 0
29 24 2 0 0 0 0
27 30 1 0 0 0 0
1 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
1 35 1 1 0 0 0
34 35 1 1 0 0 0
33 34 1 1 0 0 0
33 36 1 0 0 0 0
34 37 1 0 0 0 0
35 38 1 0 0 0 0
11 4 1 6 0 0 0
14 18 1 6 0 0 0
15 19 1 1 0 0 0
39 40 1 0 0 0 0
16 39 1 0 0 0 0
41 42 1 0 0 0 0
32 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 44
M SMT 1 ^CH2OH
M SBV 1 44 -38.3394 23.9234
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 46
M SMT 2 CH2OH
M SBV 2 46 -37.0020 24.6378
S SKP 8
ID FL1CQUCS0001
KNApSAcK_ID C00006246
NAME Safflor yellow A
CAS_RN 85532-77-0
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES C(CO)(C5O)OC(C(O)C5O)C(C=13)(O)C(O)=C(C(C=Cc(c4)ccc(O)c4)=O)C(C1C(C(O3)2)OC(CO)C(O)C(O)2)=O
M END