Mol:FL2FAEGS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.9621 0.5643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9621 0.5643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9621 -0.2386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9621 -0.2386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2669 -0.6399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2669 -0.6399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4284 -0.2386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4284 -0.2386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4284 0.5643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4284 0.5643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2669 0.9656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2669 0.9656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1236 -0.6399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1236 -0.6399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8189 -0.2386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8189 -0.2386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8189 0.5643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8189 0.5643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1236 0.9656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1236 0.9656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5136 0.9654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5136 0.9654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2283 0.5529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2283 0.5529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9428 0.9654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9428 0.9654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9428 1.7904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9428 1.7904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2283 2.2030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2283 2.2030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5136 1.7904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5136 1.7904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8189 1.5098 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8189 1.5098 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2669 -1.4422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2669 -1.4422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6569 0.9654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6569 0.9654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3676 0.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3676 0.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8303 0.6789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8303 0.6789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1842 0.0328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1842 0.0328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3902 0.4852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3902 0.4852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1227 1.1314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1227 1.1314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9166 0.6789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9166 0.6789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5923 0.0379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5923 0.0379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3469 0.1622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3469 0.1622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8325 -0.1266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8325 -0.1266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1236 -1.1562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1236 -1.1562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3834 -0.7127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3834 -0.7127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8774 -1.5683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8774 -1.5683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9273 -1.2968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9273 -1.2968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9716 -1.5683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9716 -1.5683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4775 -0.7127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4775 -0.7127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4276 -0.9841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4276 -0.9841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9716 -2.2030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9716 -2.2030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3935 -2.0221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3935 -2.0221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4695 -1.2167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4695 -1.2167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0710 -0.7127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0710 -0.7127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6557 0.5537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6557 0.5537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5412 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5412 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6483 1.4038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6483 1.4038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6557 2.2021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6557 2.2021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6483 1.6290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6483 1.6290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 9 17 1 1 0 0 0 | + | 9 17 1 1 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 20 23 1 1 0 0 0 | + | 20 23 1 1 0 0 0 |
| − | 20 24 1 0 0 0 0 | + | 20 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 21 1 0 0 0 0 | + | 25 21 1 0 0 0 0 |
| − | 23 26 1 0 0 0 0 | + | 23 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 7 29 2 0 0 0 0 | + | 7 29 2 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 33 36 1 0 0 0 0 | + | 33 36 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 30 39 1 0 0 0 0 | + | 30 39 1 0 0 0 0 |
| − | 34 26 1 0 0 0 0 | + | 34 26 1 0 0 0 0 |
| − | 13 40 1 0 0 0 0 | + | 13 40 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 25 41 1 0 0 0 0 | + | 25 41 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 14 43 1 0 0 0 0 | + | 14 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 46 0.6246 -0.4284 | + | M SBV 1 46 0.6246 -0.4284 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 48 -0.7130 -0.4116 | + | M SBV 2 48 -0.7130 -0.4116 |
| − | S SKP 5 | + | S SKP 5 |
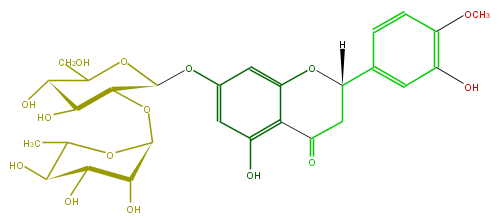
| − | ID FL2FAEGS0002 | + | ID FL2FAEGS0002 |
| − | FORMULA C28H33HO15 | + | FORMULA C28H33HO15 |
| − | EXACTMASS 610.189770418 | + | EXACTMASS 610.189770418 |
| − | AVERAGEMASS 610.56056 | + | AVERAGEMASS 610.56056 |
| − | SMILES c(c1)c(OC)c(O)cc1C([H])(O2)CC(c(c5O)c(cc(c5)OC(C3OC(O4)C(O)C(C(C4C)O)O)OC(C(C3O)O)CO)2)=O | + | SMILES c(c1)c(OC)c(O)cc1C([H])(O2)CC(c(c5O)c(cc(c5)OC(C3OC(O4)C(O)C(C(C4C)O)O)OC(C(C3O)O)CO)2)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-0.9621 0.5643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9621 -0.2386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2669 -0.6399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4284 -0.2386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4284 0.5643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2669 0.9656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1236 -0.6399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8189 -0.2386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8189 0.5643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1236 0.9656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5136 0.9654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2283 0.5529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9428 0.9654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9428 1.7904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2283 2.2030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5136 1.7904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8189 1.5098 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0
-0.2669 -1.4422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6569 0.9654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3676 0.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8303 0.6789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1842 0.0328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3902 0.4852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1227 1.1314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9166 0.6789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5923 0.0379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3469 0.1622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8325 -0.1266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1236 -1.1562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3834 -0.7127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8774 -1.5683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9273 -1.2968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9716 -1.5683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4775 -0.7127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4276 -0.9841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9716 -2.2030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3935 -2.0221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4695 -1.2167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0710 -0.7127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6557 0.5537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5412 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6483 1.4038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6557 2.2021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.6483 1.6290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
9 17 1 1 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
19 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
20 23 1 1 0 0 0
20 24 1 0 0 0 0
24 25 1 0 0 0 0
25 21 1 0 0 0 0
23 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
7 29 2 0 0 0 0
30 31 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
33 36 1 0 0 0 0
32 37 1 0 0 0 0
31 38 1 0 0 0 0
30 39 1 0 0 0 0
34 26 1 0 0 0 0
13 40 1 0 0 0 0
41 42 1 0 0 0 0
25 41 1 0 0 0 0
43 44 1 0 0 0 0
14 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 ^CH2OH
M SBV 1 46 0.6246 -0.4284
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 48
M SMT 2 OCH3
M SBV 2 48 -0.7130 -0.4116
S SKP 5
ID FL2FAEGS0002
FORMULA C28H33HO15
EXACTMASS 610.189770418
AVERAGEMASS 610.56056
SMILES c(c1)c(OC)c(O)cc1C([H])(O2)CC(c(c5O)c(cc(c5)OC(C3OC(O4)C(O)C(C(C4C)O)O)OC(C(C3O)O)CO)2)=O
M END