Mol:FL3FAACS0034
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.4050 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4050 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4050 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4050 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1513 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1513 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7076 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7076 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7076 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7076 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1513 -0.6810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1513 -0.6810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2639 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2639 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8202 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8202 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8202 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8202 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2639 -0.6810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2639 -0.6810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2639 -2.4665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2639 -2.4665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9611 -0.6811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9611 -0.6811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3760 1.7972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3760 1.7972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2295 1.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2295 1.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0274 0.6778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0274 0.6778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0342 0.0404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0342 0.0404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4976 0.5036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4976 0.5036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1925 1.0815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1925 1.0815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0920 2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0920 2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2644 2.0536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2644 2.0536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6081 0.2993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6081 0.2993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1513 -2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1513 -2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5000 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5000 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0862 -0.9164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0862 -0.9164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6724 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6724 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6724 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6724 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0862 0.4374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0862 0.4374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5000 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5000 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2583 0.4372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2583 0.4372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2499 0.6698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2499 0.6698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2499 1.1428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2499 1.1428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6677 0.4286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6677 0.4286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1513 0.7078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1513 0.7078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6276 0.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6276 0.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1715 0.7468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1715 0.7468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7153 0.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7153 0.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7153 -0.1952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7153 -0.1952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1715 -0.5092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1715 -0.5092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6276 -0.1952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6276 -0.1952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2583 -0.5087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2583 -0.5087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7678 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7678 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4823 1.0012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4823 1.0012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 9 23 1 0 0 0 0 | + | 9 23 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 21 30 1 0 0 0 0 | + | 21 30 1 0 0 0 0 |
| − | 30 31 2 0 0 0 0 | + | 30 31 2 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 18 41 1 0 0 0 0 | + | 18 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -4.8788 5.9300 | + | M SBV 1 45 -4.8788 5.9300 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAACS0034 | + | ID FL3FAACS0034 |
| − | KNApSAcK_ID C00006301 | + | KNApSAcK_ID C00006301 |
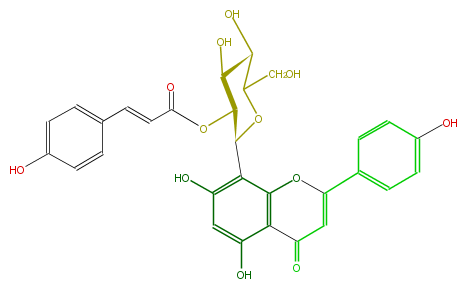
| − | NAME Vitexin 2''-O-p-coumarate | + | NAME Vitexin 2''-O-p-coumarate |
| − | CAS_RN 59282-55-2 | + | CAS_RN 59282-55-2 |
| − | FORMULA C30H26O12 | + | FORMULA C30H26O12 |
| − | EXACTMASS 578.1424262959999 | + | EXACTMASS 578.1424262959999 |
| − | AVERAGEMASS 578.5202400000001 | + | AVERAGEMASS 578.5202400000001 |
| − | SMILES C(C(O2)C(C(C(C(c(c(O)3)c(O4)c(C(C=C4c(c5)ccc(O)c5)=O)c(O)c3)2)OC(=O)C=Cc(c1)ccc(c1)O)O)O)O | + | SMILES C(C(O2)C(C(C(C(c(c(O)3)c(O4)c(C(C=C4c(c5)ccc(O)c5)=O)c(O)c3)2)OC(=O)C=Cc(c1)ccc(c1)O)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.4050 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4050 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1513 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7076 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7076 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1513 -0.6810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2639 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8202 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8202 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2639 -0.6810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2639 -2.4665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9611 -0.6811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3760 1.7972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2295 1.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0274 0.6778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0342 0.0404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4976 0.5036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1925 1.0815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0920 2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2644 2.0536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6081 0.2993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1513 -2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5000 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0862 -0.9164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6724 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6724 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0862 0.4374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5000 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2583 0.4372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2499 0.6698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2499 1.1428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6677 0.4286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1513 0.7078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6276 0.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1715 0.7468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7153 0.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7153 -0.1952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1715 -0.5092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6276 -0.1952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2583 -0.5087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7678 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4823 1.0012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
9 23 1 0 0 0 0
26 29 1 0 0 0 0
21 30 1 0 0 0 0
30 31 2 0 0 0 0
30 32 1 0 0 0 0
32 33 2 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 34 1 0 0 0 0
37 40 1 0 0 0 0
18 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -4.8788 5.9300
S SKP 8
ID FL3FAACS0034
KNApSAcK_ID C00006301
NAME Vitexin 2''-O-p-coumarate
CAS_RN 59282-55-2
FORMULA C30H26O12
EXACTMASS 578.1424262959999
AVERAGEMASS 578.5202400000001
SMILES C(C(O2)C(C(C(C(c(c(O)3)c(O4)c(C(C=C4c(c5)ccc(O)c5)=O)c(O)c3)2)OC(=O)C=Cc(c1)ccc(c1)O)O)O)O
M END