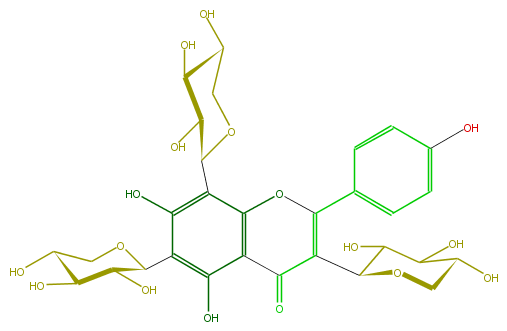
Mol:FL3FAACS0040
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 47 52 0 0 0 0 0 0 0 0999 V2000 | + | 47 52 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.6456 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6456 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6456 -1.8031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6456 -1.8031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9312 -2.2156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9312 -2.2156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2167 -1.8031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2167 -1.8031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2167 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2167 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9312 -0.5656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9312 -0.5656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4977 -2.2156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4977 -2.2156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2122 -1.8031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2122 -1.8031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2122 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2122 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4977 -0.5656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4977 -0.5656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4977 -2.8588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4977 -2.8588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3598 -0.5658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3598 -0.5658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6160 2.3458 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6160 2.3458 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3938 1.7567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3938 1.7567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0639 0.9081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0639 0.9081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0550 0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0550 0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4600 0.6844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4600 0.6844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8517 1.4266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8517 1.4266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0170 3.0403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0170 3.0403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3938 2.4108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3938 2.4108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5926 0.3794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5926 0.3794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9312 -3.0403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9312 -3.0403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0059 -0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0059 -0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7588 -0.9740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7588 -0.9740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5116 -0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5116 -0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5116 0.3301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5116 0.3301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7588 0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7588 0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0059 0.3301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0059 0.3301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2641 0.7645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2641 0.7645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0126 -1.7321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0126 -1.7321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5359 -2.3614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5359 -2.3614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8494 -2.0944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8494 -2.0944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1871 -2.0872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1871 -2.0872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6685 -1.6059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6685 -1.6059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2690 -1.9228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2690 -1.9228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6725 -2.1132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6725 -2.1132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2791 -2.3975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2791 -2.3975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1768 -2.4828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1768 -2.4828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5927 -1.6198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5927 -1.6198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0959 -2.2117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0959 -2.2117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8645 -2.1316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8645 -2.1316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5637 -2.4705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5637 -2.4705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0605 -1.8787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0605 -1.8787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2919 -1.9587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2919 -1.9587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0114 -1.6198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0114 -1.6198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9710 -1.5667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9710 -1.5667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6725 -2.2320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6725 -2.2320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 9 1 0 0 0 0 | + | 23 9 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 2 1 0 0 0 0 | + | 33 2 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 1 1 0 0 0 | + | 40 41 1 1 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 43 42 1 1 0 0 0 | + | 43 42 1 1 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 39 1 0 0 0 0 | + | 44 39 1 0 0 0 0 |
| − | 39 45 1 0 0 0 0 | + | 39 45 1 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 43 47 1 0 0 0 0 | + | 43 47 1 0 0 0 0 |
| − | 40 8 1 0 0 0 0 | + | 40 8 1 0 0 0 0 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAACS0040 | + | ID FL3FAACS0040 |
| − | FORMULA C30H34O17 | + | FORMULA C30H34O17 |
| − | EXACTMASS 666.179599662 | + | EXACTMASS 666.179599662 |
| − | AVERAGEMASS 666.58076 | + | AVERAGEMASS 666.58076 |
| − | SMILES O(C(c(c6O)c(c(c(c64)OC(c(c5)ccc(O)c5)=C(C4=O)C(C3O)OCC(O)C3O)C(O2)C(C(O)C(C2)O)O)O)1)CC(O)C(C1O)O | + | SMILES O(C(c(c6O)c(c(c(c64)OC(c(c5)ccc(O)c5)=C(C4=O)C(C3O)OCC(O)C3O)C(O2)C(C(O)C(C2)O)O)O)1)CC(O)C(C1O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
47 52 0 0 0 0 0 0 0 0999 V2000
-1.6456 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6456 -1.8031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9312 -2.2156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2167 -1.8031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2167 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9312 -0.5656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4977 -2.2156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2122 -1.8031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2122 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4977 -0.5656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4977 -2.8588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3598 -0.5658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6160 2.3458 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3938 1.7567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0639 0.9081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0550 0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4600 0.6844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8517 1.4266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0170 3.0403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3938 2.4108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5926 0.3794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9312 -3.0403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0059 -0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7588 -0.9740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5116 -0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5116 0.3301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7588 0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0059 0.3301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2641 0.7645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0126 -1.7321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5359 -2.3614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8494 -2.0944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1871 -2.0872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6685 -1.6059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2690 -1.9228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6725 -2.1132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2791 -2.3975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1768 -2.4828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5927 -1.6198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0959 -2.2117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8645 -2.1316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5637 -2.4705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0605 -1.8787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2919 -1.9587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0114 -1.6198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9710 -1.5667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6725 -2.2320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
23 9 1 0 0 0 0
26 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 2 1 0 0 0 0
39 40 1 0 0 0 0
40 41 1 1 0 0 0
41 42 1 1 0 0 0
43 42 1 1 0 0 0
43 44 1 0 0 0 0
44 39 1 0 0 0 0
39 45 1 0 0 0 0
44 46 1 0 0 0 0
43 47 1 0 0 0 0
40 8 1 0 0 0 0
S SKP 5
ID FL3FAACS0040
FORMULA C30H34O17
EXACTMASS 666.179599662
AVERAGEMASS 666.58076
SMILES O(C(c(c6O)c(c(c(c64)OC(c(c5)ccc(O)c5)=C(C4=O)C(C3O)OCC(O)C3O)C(O2)C(C(O)C(C2)O)O)O)1)CC(O)C(C1O)O
M END