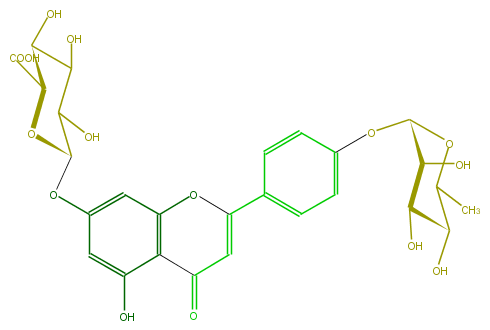
Mol:FL3FAAGS0028
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.5504 -1.0817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5504 -1.0817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5504 -1.8912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5504 -1.8912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8492 -2.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8492 -2.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1481 -1.8912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1481 -1.8912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1481 -1.0817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1481 -1.0817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8492 -0.6769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8492 -0.6769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4470 -2.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4470 -2.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2541 -1.8912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2541 -1.8912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2541 -1.0817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2541 -1.0817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4470 -0.6769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4470 -0.6769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4456 -3.0935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4456 -3.0935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9550 -0.6770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9550 -0.6770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6695 -1.0895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6695 -1.0895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3840 -0.6770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3840 -0.6770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3840 0.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3840 0.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6695 0.5606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6695 0.5606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9550 0.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9550 0.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2512 -0.6770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2512 -0.6770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8492 -3.1052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8492 -3.1052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0985 0.5605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0985 0.5605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7031 0.5363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7031 0.5363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9113 0.0790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9113 0.0790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1625 0.9581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1625 0.9581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9113 1.8426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9113 1.8426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7031 2.2998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7031 2.2998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4519 1.4207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4519 1.4207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3204 2.9479 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3204 2.9479 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9100 2.4529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9100 2.4529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5629 0.4875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5629 0.4875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8636 -0.9542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8636 -0.9542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6555 -1.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6555 -1.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4042 -0.5322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4042 -0.5322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6555 0.3521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6555 0.3521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8636 0.8094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8636 0.8094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1148 -0.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1148 -0.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8671 -0.0697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8671 -0.0697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9062 -1.6870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9062 -1.6870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4073 -2.1942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4073 -2.1942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9746 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9746 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0743 2.0655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0743 2.0655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0743 3.1052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0743 3.1052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9746 1.5456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9746 1.5456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 18 22 1 0 0 0 0 | + | 18 22 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 30 1 1 0 0 0 | + | 35 30 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 30 37 1 0 0 0 0 | + | 30 37 1 0 0 0 0 |
| − | 34 20 1 0 0 0 0 | + | 34 20 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 40 42 1 0 0 0 0 | + | 40 42 1 0 0 0 0 |
| − | 26 40 1 0 0 0 0 | + | 26 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 40 41 42 | + | M SAL 1 3 40 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^COOH | + | M SMT 1 ^COOH |
| − | M SBV 1 46 0.6224 -0.6448 | + | M SBV 1 46 0.6224 -0.6448 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAAGS0028 | + | ID FL3FAAGS0028 |
| − | FORMULA C27H28O15 | + | FORMULA C27H28O15 |
| − | EXACTMASS 592.1428202259999 | + | EXACTMASS 592.1428202259999 |
| − | AVERAGEMASS 592.5022200000001 | + | AVERAGEMASS 592.5022200000001 |
| − | SMILES c(c1OC(O5)C(C(C(O)C5C(O)=O)O)O)c(O)c(C4=O)c(OC(=C4)c(c3)ccc(c3)OC(O2)C(O)C(C(C(C)2)O)O)c1 | + | SMILES c(c1OC(O5)C(C(C(O)C5C(O)=O)O)O)c(O)c(C4=O)c(OC(=C4)c(c3)ccc(c3)OC(O2)C(O)C(C(C(C)2)O)O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-2.5504 -1.0817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5504 -1.8912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8492 -2.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1481 -1.8912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1481 -1.0817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8492 -0.6769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4470 -2.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2541 -1.8912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2541 -1.0817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4470 -0.6769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4456 -3.0935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9550 -0.6770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6695 -1.0895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3840 -0.6770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3840 0.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6695 0.5606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9550 0.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2512 -0.6770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8492 -3.1052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0985 0.5605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7031 0.5363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9113 0.0790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1625 0.9581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9113 1.8426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7031 2.2998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4519 1.4207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3204 2.9479 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9100 2.4529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5629 0.4875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8636 -0.9542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6555 -1.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4042 -0.5322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6555 0.3521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8636 0.8094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1148 -0.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8671 -0.0697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9062 -1.6870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4073 -2.1942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9746 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0743 2.0655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0743 3.1052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9746 1.5456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
18 22 1 0 0 0 0
23 29 1 0 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 30 1 1 0 0 0
35 36 1 0 0 0 0
30 37 1 0 0 0 0
34 20 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
40 41 2 0 0 0 0
40 42 1 0 0 0 0
26 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 40 41 42
M SBL 1 1 46
M SMT 1 ^COOH
M SBV 1 46 0.6224 -0.6448
S SKP 5
ID FL3FAAGS0028
FORMULA C27H28O15
EXACTMASS 592.1428202259999
AVERAGEMASS 592.5022200000001
SMILES c(c1OC(O5)C(C(C(O)C5C(O)=O)O)O)c(O)c(C4=O)c(OC(=C4)c(c3)ccc(c3)OC(O2)C(O)C(C(C(C)2)O)O)c1
M END