Mol:FL3FACCS0043
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.0799 -0.8433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0799 -0.8433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0799 -1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0799 -1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4764 -1.8068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4764 -1.8068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0327 -1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0327 -1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0327 -0.8433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0327 -0.8433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4764 -0.5221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4764 -0.5221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5890 -1.8068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5890 -1.8068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1453 -1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1453 -1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1453 -0.8433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1453 -0.8433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5890 -0.5221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5890 -0.5221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5890 -2.3076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5890 -2.3076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6360 -0.5222 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6360 -0.5222 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4764 -2.4489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4764 -2.4489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7633 -0.5016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7633 -0.5016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3495 -0.8400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3495 -0.8400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9357 -0.5016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9357 -0.5016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9357 0.1753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9357 0.1753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3495 0.5138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3495 0.5138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7633 0.1753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7633 0.1753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5216 0.5136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5216 0.5136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8249 1.8118 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 0.8249 1.8118 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 0.2193 1.3530 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.2193 1.3530 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.4762 0.6923 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.4762 0.6923 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.4831 0.0549 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.4831 0.0549 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.9464 0.5181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9464 0.5181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6414 1.0961 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.6414 1.0961 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.4570 2.4489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4570 2.4489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1845 2.0682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1845 2.0682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1592 0.3138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1592 0.3138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1873 -0.8405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1873 -0.8405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1873 -1.4757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1873 -1.4757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7374 -0.5228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7374 -0.5228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2863 -0.8398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2863 -0.8398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8353 -0.5228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8353 -0.5228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8353 0.1262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8353 0.1262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3974 0.4508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3974 0.4508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9595 0.1262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9595 0.1262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9595 -0.5228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9595 -0.5228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3974 -0.8474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3974 -0.8474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5216 0.4507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5216 0.4507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5216 -0.8473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5216 -0.8473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5216 -0.8398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5216 -0.8398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4383 1.0961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4383 1.0961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9383 1.9621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9383 1.9621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 3 13 1 0 0 0 0 | + | 3 13 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 14 9 1 0 0 0 0 | + | 14 9 1 0 0 0 0 |
| − | 17 20 1 0 0 0 0 | + | 17 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 6 24 1 0 0 0 0 | + | 6 24 1 0 0 0 0 |
| − | 12 30 1 0 0 0 0 | + | 12 30 1 0 0 0 0 |
| − | 30 31 2 0 0 0 0 | + | 30 31 2 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 16 42 1 0 0 0 0 | + | 16 42 1 0 0 0 0 |
| − | 26 43 1 0 0 0 0 | + | 26 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 47 1.4383 1.0961 | + | M SVB 1 47 1.4383 1.0961 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACCS0043 | + | ID FL3FACCS0043 |
| − | KNApSAcK_ID C00006379 | + | KNApSAcK_ID C00006379 |
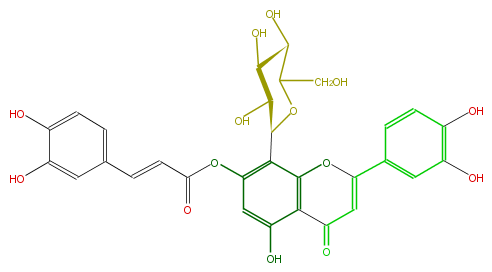
| − | NAME Orientin 7-O-caffeate | + | NAME Orientin 7-O-caffeate |
| − | CAS_RN 156298-99-6 | + | CAS_RN 156298-99-6 |
| − | FORMULA C30H26O14 | + | FORMULA C30H26O14 |
| − | EXACTMASS 610.13225554 | + | EXACTMASS 610.13225554 |
| − | AVERAGEMASS 610.51904 | + | AVERAGEMASS 610.51904 |
| − | SMILES c(c1)(O)c(ccc(C(=C4)Oc(c([C@H](O5)[C@@H](O)[C@H]([C@@H](O)C5CO)O)2)c(C4=O)c(cc2OC(=O)C=Cc(c3)ccc(O)c(O)3)O)1)O | + | SMILES c(c1)(O)c(ccc(C(=C4)Oc(c([C@H](O5)[C@@H](O)[C@H]([C@@H](O)C5CO)O)2)c(C4=O)c(cc2OC(=O)C=Cc(c3)ccc(O)c(O)3)O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-0.0799 -0.8433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0799 -1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4764 -1.8068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0327 -1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0327 -0.8433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4764 -0.5221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5890 -1.8068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1453 -1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1453 -0.8433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5890 -0.5221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5890 -2.3076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6360 -0.5222 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4764 -2.4489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7633 -0.5016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3495 -0.8400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9357 -0.5016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9357 0.1753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3495 0.5138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7633 0.1753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5216 0.5136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8249 1.8118 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
0.2193 1.3530 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.4762 0.6923 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.4831 0.0549 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.9464 0.5181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6414 1.0961 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.4570 2.4489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1845 2.0682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1592 0.3138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1873 -0.8405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1873 -1.4757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7374 -0.5228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2863 -0.8398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8353 -0.5228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8353 0.1262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3974 0.4508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9595 0.1262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9595 -0.5228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3974 -0.8474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5216 0.4507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5216 -0.8473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5216 -0.8398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4383 1.0961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9383 1.9621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
3 13 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
14 9 1 0 0 0 0
17 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
6 24 1 0 0 0 0
12 30 1 0 0 0 0
30 31 2 0 0 0 0
30 32 1 0 0 0 0
32 33 2 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 34 1 0 0 0 0
37 40 1 0 0 0 0
38 41 1 0 0 0 0
16 42 1 0 0 0 0
26 43 1 0 0 0 0
43 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 47
M SMT 1 CH2OH
M SVB 1 47 1.4383 1.0961
S SKP 8
ID FL3FACCS0043
KNApSAcK_ID C00006379
NAME Orientin 7-O-caffeate
CAS_RN 156298-99-6
FORMULA C30H26O14
EXACTMASS 610.13225554
AVERAGEMASS 610.51904
SMILES c(c1)(O)c(ccc(C(=C4)Oc(c([C@H](O5)[C@@H](O)[C@H]([C@@H](O)C5CO)O)2)c(C4=O)c(cc2OC(=O)C=Cc(c3)ccc(O)c(O)3)O)1)O
M END