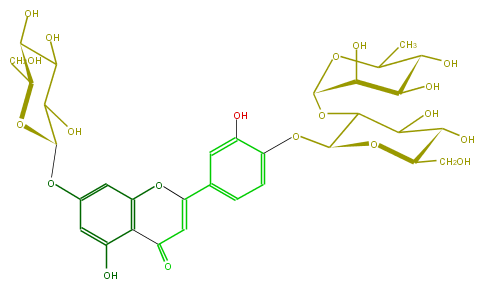
Mol:FL3FACGS0046
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.1160 -1.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1160 -1.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1160 -2.0189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1160 -2.0189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5098 -2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5098 -2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9036 -2.0189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9036 -2.0189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9036 -1.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9036 -1.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5098 -0.9689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5098 -0.9689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2975 -2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2975 -2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6913 -2.0189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6913 -2.0189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6913 -1.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6913 -1.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2975 -0.9689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2975 -0.9689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0668 -2.8632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0668 -2.8632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0854 -0.9691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0854 -0.9691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5324 -1.3257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5324 -1.3257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1501 -0.9691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1501 -0.9691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1501 -0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1501 -0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5324 0.1010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5324 0.1010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0854 -0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0854 -0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8140 -0.9158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8140 -0.9158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5098 -3.0672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5098 -3.0672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9191 0.1837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9191 0.1837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5391 0.4548 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5391 0.4548 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6949 -0.0327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6949 -0.0327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9627 0.9047 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9627 0.9047 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6949 1.8478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6949 1.8478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5391 2.3353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5391 2.3353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2712 1.3979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2712 1.3979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3430 3.0672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3430 3.0672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8693 2.4987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8693 2.4987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3527 0.2947 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3527 0.2947 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3799 0.6930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3799 0.6930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7101 -0.1052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7101 -0.1052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7464 0.0028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7464 0.0028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6893 -0.4542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6893 -0.4542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3590 0.3440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3590 0.3440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3227 0.2361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3227 0.2361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8450 2.0569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8450 2.0569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3240 1.1546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3240 1.1546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3258 1.4409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3258 1.4409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3335 1.1546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3335 1.1546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8547 2.0569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8547 2.0569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8528 1.7706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8528 1.7706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4047 2.3225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4047 2.3225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4535 1.8964 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4535 1.8964 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0431 1.3447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0431 1.3447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5378 0.6787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5378 0.6787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3258 2.3137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3258 2.3137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0026 0.7121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0026 0.7121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8525 0.1139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8525 0.1139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5324 0.6642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5324 0.6642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8281 1.9547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8281 1.9547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8525 1.0848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8525 1.0848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3384 -0.3974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3384 -0.3974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1459 -1.4892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1459 -1.4892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 22 18 1 0 0 0 0 | + | 22 18 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 1 0 0 0 | + | 37 38 1 1 0 0 0 |
| − | 38 39 1 1 0 0 0 | + | 38 39 1 1 0 0 0 |
| − | 40 39 1 1 0 0 0 | + | 40 39 1 1 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | 39 44 1 0 0 0 0 | + | 39 44 1 0 0 0 0 |
| − | 31 20 1 0 0 0 0 | + | 31 20 1 0 0 0 0 |
| − | 30 45 1 0 0 0 0 | + | 30 45 1 0 0 0 0 |
| − | 37 45 1 0 0 0 0 | + | 37 45 1 0 0 0 0 |
| − | 38 46 1 0 0 0 0 | + | 38 46 1 0 0 0 0 |
| − | 35 47 1 0 0 0 0 | + | 35 47 1 0 0 0 0 |
| − | 34 48 1 0 0 0 0 | + | 34 48 1 0 0 0 0 |
| − | 16 49 1 0 0 0 0 | + | 16 49 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 26 50 1 0 0 0 0 | + | 26 50 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 33 52 1 0 0 0 0 | + | 33 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 56 0.5568 -0.5568 | + | M SBV 1 56 0.5568 -0.5568 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 58 -0.6492 -0.0568 | + | M SBV 2 58 -0.6492 -0.0568 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FACGS0046 | + | ID FL3FACGS0046 |
| − | FORMULA C33H40O20 | + | FORMULA C33H40O20 |
| − | EXACTMASS 756.21129372 | + | EXACTMASS 756.21129372 |
| − | AVERAGEMASS 756.6587 | + | AVERAGEMASS 756.6587 |
| − | SMILES C(C1OC(O6)C(C(C(C(C)6)O)O)O)(O)C(O)C(OC(Oc(c2)c(O)cc(C(O3)=CC(c(c(O)4)c3cc(OC(O5)C(C(O)C(O)C5CO)O)c4)=O)c2)1)CO | + | SMILES C(C1OC(O6)C(C(C(C(C)6)O)O)O)(O)C(O)C(OC(Oc(c2)c(O)cc(C(O3)=CC(c(c(O)4)c3cc(OC(O5)C(C(O)C(O)C5CO)O)c4)=O)c2)1)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-3.1160 -1.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1160 -2.0189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5098 -2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9036 -2.0189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9036 -1.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5098 -0.9689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2975 -2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6913 -2.0189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6913 -1.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2975 -0.9689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0668 -2.8632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0854 -0.9691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5324 -1.3257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1501 -0.9691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1501 -0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5324 0.1010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0854 -0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8140 -0.9158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5098 -3.0672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9191 0.1837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5391 0.4548 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6949 -0.0327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9627 0.9047 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6949 1.8478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5391 2.3353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2712 1.3979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3430 3.0672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8693 2.4987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3527 0.2947 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3799 0.6930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7101 -0.1052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7464 0.0028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6893 -0.4542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3590 0.3440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3227 0.2361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8450 2.0569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3240 1.1546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3258 1.4409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3335 1.1546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8547 2.0569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8528 1.7706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4047 2.3225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4535 1.8964 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0431 1.3447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5378 0.6787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3258 2.3137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0026 0.7121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.8525 0.1139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5324 0.6642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8281 1.9547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.8525 1.0848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3384 -0.3974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1459 -1.4892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 1 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
23 29 1 0 0 0 0
22 18 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
36 37 1 0 0 0 0
37 38 1 1 0 0 0
38 39 1 1 0 0 0
40 39 1 1 0 0 0
40 41 1 0 0 0 0
41 36 1 0 0 0 0
41 42 1 0 0 0 0
40 43 1 0 0 0 0
39 44 1 0 0 0 0
31 20 1 0 0 0 0
30 45 1 0 0 0 0
37 45 1 0 0 0 0
38 46 1 0 0 0 0
35 47 1 0 0 0 0
34 48 1 0 0 0 0
16 49 1 0 0 0 0
50 51 1 0 0 0 0
26 50 1 0 0 0 0
52 53 1 0 0 0 0
33 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 56
M SMT 1 ^ CH2OH
M SBV 1 56 0.5568 -0.5568
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 58
M SMT 2 CH2OH
M SBV 2 58 -0.6492 -0.0568
S SKP 5
ID FL3FACGS0046
FORMULA C33H40O20
EXACTMASS 756.21129372
AVERAGEMASS 756.6587
SMILES C(C1OC(O6)C(C(C(C(C)6)O)O)O)(O)C(O)C(OC(Oc(c2)c(O)cc(C(O3)=CC(c(c(O)4)c3cc(OC(O5)C(C(O)C(O)C5CO)O)c4)=O)c2)1)CO
M END