Mol:FL3FACGS0047
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.3507 -1.3164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3507 -1.3164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3507 -1.8373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3507 -1.8373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1004 -2.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1004 -2.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5514 -1.8373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5514 -1.8373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5514 -1.3164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5514 -1.3164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1004 -1.0560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1004 -1.0560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0025 -2.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0025 -2.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4535 -1.8373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4535 -1.8373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4535 -1.3164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4535 -1.3164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0025 -1.0560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0025 -1.0560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1742 -2.4656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1742 -2.4656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9044 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9044 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3642 -1.3215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3642 -1.3215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8239 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8239 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8239 -0.5253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8239 -0.5253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3642 -0.2599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3642 -0.2599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9044 -0.5253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9044 -0.5253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8702 -1.0165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8702 -1.0165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1004 -2.6175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1004 -2.6175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4182 -0.1821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4182 -0.1821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2835 -1.3215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2835 -1.3215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0822 0.8174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0822 0.8174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7104 0.4547 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.7104 0.4547 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.5111 1.1522 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.5111 1.1522 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 3.7104 1.8539 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.7104 1.8539 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 3.0822 2.2167 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.0822 2.2167 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.2815 1.5191 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 3.2815 1.5191 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 2.7988 2.7076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7988 2.7076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1024 2.0802 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1024 2.0802 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9731 0.8855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9731 0.8855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3966 -0.8612 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.3966 -0.8612 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.8809 -1.5418 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.8809 -1.5418 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1384 -1.2531 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1384 -1.2531 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.4220 -1.2453 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.4220 -1.2453 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.9426 -0.7246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9426 -0.7246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7011 -0.9969 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.7011 -0.9969 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.9731 -1.1940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9731 -1.1940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6847 -1.5809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6847 -1.5809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7130 -1.9672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7130 -1.9672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1342 -0.2132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1342 -0.2132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9965 1.1481 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -1.9965 1.1481 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -1.4809 0.4674 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.4809 0.4674 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.7384 0.7562 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.7384 0.7562 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.0219 0.7639 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.0219 0.7639 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.5425 1.2847 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.5425 1.2847 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -1.1921 0.9418 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1921 0.9418 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7103 0.7360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7103 0.7360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2847 0.4283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2847 0.4283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3129 0.0420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3129 0.0420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9781 0.7639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9781 0.7639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5123 1.7941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5123 1.7941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7500 1.1469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7500 1.1469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0251 1.3262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0251 1.3262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4606 2.2264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4606 2.2264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 14 21 1 0 0 0 0 | + | 14 21 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 20 23 1 0 0 0 0 | + | 20 23 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 44 43 1 1 0 0 0 | + | 44 43 1 1 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 47 40 1 0 0 0 0 | + | 47 40 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 34 18 1 0 0 0 0 | + | 34 18 1 0 0 0 0 |
| − | 27 51 1 0 0 0 0 | + | 27 51 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 45 53 1 0 0 0 0 | + | 45 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 53 54 | + | M SAL 2 2 53 54 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 58 0.0251 1.3262 | + | M SVB 2 58 0.0251 1.3262 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 51 52 | + | M SAL 1 2 51 52 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 56 2.7006 2.0934 | + | M SVB 1 56 2.7006 2.0934 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACGS0047 | + | ID FL3FACGS0047 |
| − | KNApSAcK_ID C00004307 | + | KNApSAcK_ID C00004307 |
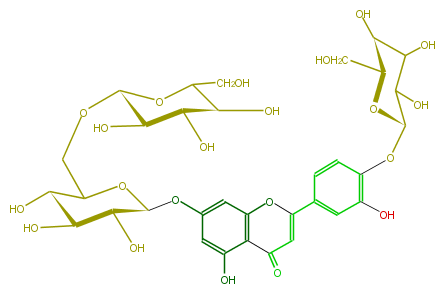
| − | NAME Luteolin 7-gentiobioside-4'-glucoside | + | NAME Luteolin 7-gentiobioside-4'-glucoside |
| − | CAS_RN 89718-43-4 | + | CAS_RN 89718-43-4 |
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
| − | SMILES O[C@@H]([C@@H](O)1)[C@H](Oc(c6)cc(O)c(c56)C(=O)C=C(O5)c(c3)cc(c(O[C@@H](O4)C(C([C@@H]([C@@H]4CO)O)O)O)c3)O)OC(CO[C@H]([C@@H]2O)OC(CO)[C@@H](O)[C@H]2O)[C@@H]1O | + | SMILES O[C@@H]([C@@H](O)1)[C@H](Oc(c6)cc(O)c(c56)C(=O)C=C(O5)c(c3)cc(c(O[C@@H](O4)C(C([C@@H]([C@@H]4CO)O)O)O)c3)O)OC(CO[C@H]([C@@H]2O)OC(CO)[C@@H](O)[C@H]2O)[C@@H]1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-0.3507 -1.3164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3507 -1.8373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1004 -2.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5514 -1.8373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5514 -1.3164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1004 -1.0560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0025 -2.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4535 -1.8373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4535 -1.3164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0025 -1.0560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1742 -2.4656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9044 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3642 -1.3215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8239 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8239 -0.5253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3642 -0.2599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9044 -0.5253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8702 -1.0165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1004 -2.6175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4182 -0.1821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2835 -1.3215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0822 0.8174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7104 0.4547 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.5111 1.1522 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.7104 1.8539 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.0822 2.2167 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.2815 1.5191 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.7988 2.7076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1024 2.0802 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9731 0.8855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3966 -0.8612 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.8809 -1.5418 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.1384 -1.2531 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.4220 -1.2453 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.9426 -0.7246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7011 -0.9969 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.9731 -1.1940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6847 -1.5809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7130 -1.9672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1342 -0.2132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9965 1.1481 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.4809 0.4674 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.7384 0.7562 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.0219 0.7639 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.5425 1.2847 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-1.1921 0.9418 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7103 0.7360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2847 0.4283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3129 0.0420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9781 0.7639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5123 1.7941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7500 1.1469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0251 1.3262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4606 2.2264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 1 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
14 21 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
20 23 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
36 40 1 0 0 0 0
41 42 1 1 0 0 0
42 43 1 1 0 0 0
44 43 1 1 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
46 41 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
47 40 1 0 0 0 0
44 50 1 0 0 0 0
34 18 1 0 0 0 0
27 51 1 0 0 0 0
51 52 1 0 0 0 0
45 53 1 0 0 0 0
53 54 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 53 54
M SBL 2 1 58
M SMT 2 CH2OH
M SVB 2 58 0.0251 1.3262
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 51 52
M SBL 1 1 56
M SMT 1 CH2OH
M SVB 1 56 2.7006 2.0934
S SKP 8
ID FL3FACGS0047
KNApSAcK_ID C00004307
NAME Luteolin 7-gentiobioside-4'-glucoside
CAS_RN 89718-43-4
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES O[C@@H]([C@@H](O)1)[C@H](Oc(c6)cc(O)c(c56)C(=O)C=C(O5)c(c3)cc(c(O[C@@H](O4)C(C([C@@H]([C@@H]4CO)O)O)O)c3)O)OC(CO[C@H]([C@@H]2O)OC(CO)[C@@H](O)[C@H]2O)[C@@H]1O
M END