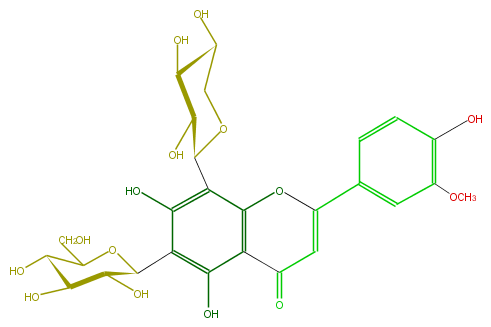
Mol:FL3FADCS0009
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4536 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4536 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4536 -1.7668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4536 -1.7668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7391 -2.1793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7391 -2.1793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0247 -1.7668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0247 -1.7668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0247 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0247 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7391 -0.5295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7391 -0.5295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6898 -2.1793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6898 -2.1793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4042 -1.7668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4042 -1.7668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4042 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4042 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6898 -0.5295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6898 -0.5295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6898 -2.8225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6898 -2.8225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1677 -0.5296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1677 -0.5296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5743 2.3645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5743 2.3645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3520 1.7753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3520 1.7753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0220 0.9268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0220 0.9268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0132 0.1082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0132 0.1082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4181 0.7031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4181 0.7031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8099 1.4454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8099 1.4454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9434 3.0040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9434 3.0040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3520 2.4866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3520 2.4866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4506 0.1844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4506 0.1844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7391 -3.0040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7391 -3.0040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2772 -0.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2772 -0.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0301 -0.8318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0301 -0.8318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7829 -0.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7829 -0.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7829 0.4722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7829 0.4722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0301 0.9068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0301 0.9068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2772 0.4722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2772 0.4722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5353 0.9066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5353 0.9066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9792 -1.8492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9792 -1.8492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5025 -2.4783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5025 -2.4783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8161 -2.2114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8161 -2.2114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1538 -2.2043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1538 -2.2043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6350 -1.7228 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6350 -1.7228 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2355 -2.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2355 -2.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4824 -2.1397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4824 -2.1397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1932 -2.6634 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1932 -2.6634 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1403 -2.6016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1403 -2.6016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1718 -0.6632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1718 -0.6632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9833 -1.3442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9833 -1.3442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7563 -1.5190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7563 -1.5190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9833 -1.1395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9833 -1.1395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 9 23 1 0 0 0 0 | + | 9 23 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 2 1 0 0 0 0 | + | 33 2 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 25 39 1 0 0 0 0 | + | 25 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 44 -0.3889 0.2661 | + | M SBV 1 44 -0.3889 0.2661 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 46 0.5208 -0.5208 | + | M SBV 2 46 0.5208 -0.5208 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FADCS0009 | + | ID FL3FADCS0009 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES C(C(C1O)OC(c(c4O)c(c(c(c4C(O5)C(O)C(O)C(O)C5)2)C(=O)C=C(c(c3)cc(OC)c(O)c3)O2)O)C(C1O)O)O | + | SMILES C(C(C1O)OC(c(c4O)c(c(c(c4C(O5)C(O)C(O)C(O)C5)2)C(=O)C=C(c(c3)cc(OC)c(O)c3)O2)O)C(C1O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-1.4536 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4536 -1.7668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7391 -2.1793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0247 -1.7668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0247 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7391 -0.5295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6898 -2.1793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4042 -1.7668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4042 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6898 -0.5295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6898 -2.8225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1677 -0.5296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5743 2.3645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3520 1.7753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0220 0.9268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0132 0.1082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4181 0.7031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8099 1.4454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9434 3.0040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3520 2.4866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4506 0.1844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7391 -3.0040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2772 -0.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0301 -0.8318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7829 -0.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7829 0.4722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0301 0.9068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2772 0.4722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5353 0.9066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9792 -1.8492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5025 -2.4783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8161 -2.2114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1538 -2.2043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6350 -1.7228 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2355 -2.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4824 -2.1397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1932 -2.6634 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1403 -2.6016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1718 -0.6632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9833 -1.3442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7563 -1.5190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9833 -1.1395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
9 23 1 0 0 0 0
26 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 2 1 0 0 0 0
39 40 1 0 0 0 0
25 39 1 0 0 0 0
41 42 1 0 0 0 0
35 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 44
M SMT 1 OCH3
M SBV 1 44 -0.3889 0.2661
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 46
M SMT 2 ^ CH2OH
M SBV 2 46 0.5208 -0.5208
S SKP 5
ID FL3FADCS0009
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES C(C(C1O)OC(c(c4O)c(c(c(c4C(O5)C(O)C(O)C(O)C5)2)C(=O)C=C(c(c3)cc(OC)c(O)c3)O2)O)C(C1O)O)O
M END