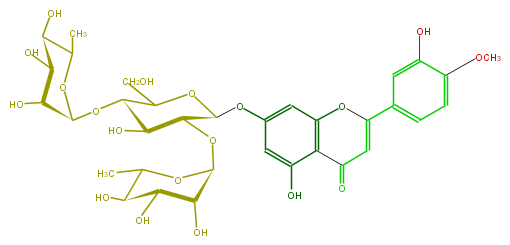
Mol:FL3FAEGS0009
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.1207 0.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1207 0.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1207 -0.7258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1207 -0.7258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5500 -1.1130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5500 -1.1130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2208 -0.7258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2208 -0.7258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2208 0.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2208 0.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5500 0.4361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5500 0.4361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8917 -1.1130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8917 -1.1130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5624 -0.7258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5624 -0.7258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5624 0.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5624 0.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8917 0.4361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8917 0.4361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8917 -1.7170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8917 -1.7170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2329 0.4360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2329 0.4360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9166 0.0411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9166 0.0411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6003 0.4360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6003 0.4360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6003 1.2253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6003 1.2253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9166 1.6200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9166 1.6200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2329 1.2253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2329 1.2253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9166 2.4094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9166 2.4094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7902 0.4352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7902 0.4352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5500 -1.8860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5500 -1.8860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8550 0.6017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8550 0.6017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2159 -0.2419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2159 -0.2419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2958 0.1160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2958 0.1160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4080 0.1256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4080 0.1256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0531 0.7708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0531 0.7708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9931 0.4334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9931 0.4334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5475 0.3115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5475 0.3115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9426 -0.1886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9426 -0.1886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4991 -0.4405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4991 -0.4405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3361 -1.2195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3361 -1.2195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8165 -2.0516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8165 -2.0516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8926 -1.7876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8926 -1.7876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9633 -2.0516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9633 -2.0516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4827 -1.2195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4827 -1.2195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4067 -1.4835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4067 -1.4835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1444 -1.3156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1444 -1.3156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2905 -1.9337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2905 -1.9337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4359 -2.5330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4359 -2.5330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9206 -2.8637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9206 -2.8637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9474 0.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9474 0.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1689 0.0638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1689 0.0638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4159 0.9282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4159 0.9282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1689 1.7978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1689 1.7978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9474 2.2474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9474 2.2474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7004 1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7004 1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7354 2.8637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7354 2.8637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1689 2.3588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1689 2.3588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.3375 1.8580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.3375 1.8580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.5404 0.4787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.5404 0.4787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7911 1.1026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7911 1.1026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9056 0.5914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9056 0.5914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4780 1.7321 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4780 1.7321 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5404 1.1187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5404 1.1187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 16 18 1 0 0 0 0 | + | 16 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 41 40 1 1 0 0 0 | + | 41 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 45 40 1 1 0 0 0 | + | 45 40 1 1 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 43 47 1 0 0 0 0 | + | 43 47 1 0 0 0 0 |
| − | 45 48 1 0 0 0 0 | + | 45 48 1 0 0 0 0 |
| − | 40 49 1 0 0 0 0 | + | 40 49 1 0 0 0 0 |
| − | 41 27 1 0 0 0 0 | + | 41 27 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 26 50 1 0 0 0 0 | + | 26 50 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 15 52 1 0 0 0 0 | + | 15 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 56 0.7980 -0.6692 | + | M SBV 1 56 0.7980 -0.6692 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 58 -0.8777 -0.5068 | + | M SBV 2 58 -0.8777 -0.5068 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAEGS0009 | + | ID FL3FAEGS0009 |
| − | FORMULA C34H42O19 | + | FORMULA C34H42O19 |
| − | EXACTMASS 754.2320291619999 | + | EXACTMASS 754.2320291619999 |
| − | AVERAGEMASS 754.68588 | + | AVERAGEMASS 754.68588 |
| − | SMILES c(c(O)6)(c1cc(c6)OC(C4OC(C(O)5)OC(C(C5O)O)C)OC(CO)C(C4O)OC(C(O)3)OC(C(C3O)O)C)C(=O)C=C(c(c2)cc(c(OC)c2)O)O1 | + | SMILES c(c(O)6)(c1cc(c6)OC(C4OC(C(O)5)OC(C(C5O)O)C)OC(CO)C(C4O)OC(C(O)3)OC(C(C3O)O)C)C(=O)C=C(c(c2)cc(c(OC)c2)O)O1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-0.1207 0.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1207 -0.7258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5500 -1.1130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2208 -0.7258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2208 0.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5500 0.4361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8917 -1.1130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5624 -0.7258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5624 0.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8917 0.4361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8917 -1.7170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2329 0.4360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9166 0.0411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6003 0.4360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6003 1.2253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9166 1.6200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2329 1.2253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9166 2.4094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7902 0.4352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5500 -1.8860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8550 0.6017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2159 -0.2419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2958 0.1160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4080 0.1256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0531 0.7708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9931 0.4334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5475 0.3115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9426 -0.1886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4991 -0.4405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3361 -1.2195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8165 -2.0516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8926 -1.7876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9633 -2.0516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4827 -1.2195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4067 -1.4835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1444 -1.3156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2905 -1.9337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4359 -2.5330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9206 -2.8637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.9474 0.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1689 0.0638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4159 0.9282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1689 1.7978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9474 2.2474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7004 1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7354 2.8637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1689 2.3588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.3375 1.8580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.5404 0.4787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7911 1.1026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9056 0.5914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4780 1.7321 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.5404 1.1187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
16 18 1 0 0 0 0
1 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 29 1 0 0 0 0
24 19 1 0 0 0 0
41 40 1 1 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 1 0 0 0
45 40 1 1 0 0 0
44 46 1 0 0 0 0
43 47 1 0 0 0 0
45 48 1 0 0 0 0
40 49 1 0 0 0 0
41 27 1 0 0 0 0
50 51 1 0 0 0 0
26 50 1 0 0 0 0
52 53 1 0 0 0 0
15 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 56
M SMT 1 ^CH2OH
M SBV 1 56 0.7980 -0.6692
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 58
M SMT 2 OCH3
M SBV 2 58 -0.8777 -0.5068
S SKP 5
ID FL3FAEGS0009
FORMULA C34H42O19
EXACTMASS 754.2320291619999
AVERAGEMASS 754.68588
SMILES c(c(O)6)(c1cc(c6)OC(C4OC(C(O)5)OC(C(C5O)O)C)OC(CO)C(C4O)OC(C(O)3)OC(C(C3O)O)C)C(=O)C=C(c(c2)cc(c(OC)c2)O)O1
M END