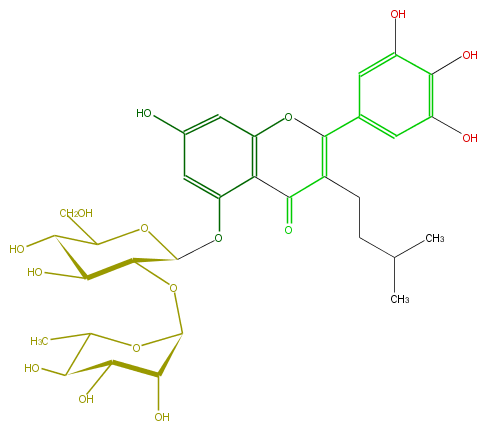
Mol:FL3FAGGI0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 48 52 0 0 0 0 0 0 0 0999 V2000 | + | 48 52 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1905 1.6266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1905 1.6266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1905 0.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1905 0.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4897 0.3484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4897 0.3484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2114 0.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2114 0.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2114 1.5625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2114 1.5625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4897 1.9671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4897 1.9671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9124 0.3484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9124 0.3484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6133 0.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6133 0.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6133 1.5625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6133 1.5625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9124 1.9671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9124 1.9671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9124 -0.2827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9124 -0.2827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3141 1.9671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3141 1.9671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0286 1.5545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0286 1.5545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7430 1.9671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7430 1.9671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7430 2.7920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7430 2.7920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0286 3.2045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0286 3.2045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3141 2.7920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3141 2.7920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4897 -0.4594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4897 -0.4594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4573 3.2045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4573 3.2045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4573 1.5547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4573 1.5547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9074 2.0405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9074 2.0405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0286 4.0294 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0286 4.0294 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3130 0.3492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3130 0.3492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3130 -0.4585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3130 -0.4585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0125 -0.8624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0125 -0.8624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7119 -0.4585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7119 -0.4585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0125 -1.6702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0125 -1.6702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7832 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7832 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1421 -1.2755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1421 -1.2755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2190 -0.9165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2190 -0.9165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3282 -0.9069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3282 -0.9069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9756 -0.2594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9756 -0.2594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9185 -0.5981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9185 -0.5981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4573 -0.6795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4573 -0.6795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0988 -1.1399 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0988 -1.1399 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4000 -1.4552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4000 -1.4552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0771 -2.3813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0771 -2.3813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5592 -3.2161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5592 -3.2161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6323 -2.9512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6323 -2.9512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6999 -3.2161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6999 -3.2161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2179 -2.3813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2179 -2.3813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1447 -2.6461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1447 -2.6461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9709 -2.5194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9709 -2.5194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1602 -3.0550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1602 -3.0550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2182 -3.6783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2182 -3.6783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6999 -4.0294 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6999 -4.0294 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5984 0.0460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5984 0.0460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7101 -0.4669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7101 -0.4669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 21 1 1 0 0 0 0 | + | 21 1 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 8 23 1 0 0 0 0 | + | 8 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 1 0 0 0 | + | 38 39 1 1 0 0 0 |
| − | 39 40 1 1 0 0 0 | + | 39 40 1 1 0 0 0 |
| − | 41 40 1 1 0 0 0 | + | 41 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 37 1 0 0 0 0 | + | 42 37 1 0 0 0 0 |
| − | 37 43 1 0 0 0 0 | + | 37 43 1 0 0 0 0 |
| − | 38 44 1 0 0 0 0 | + | 38 44 1 0 0 0 0 |
| − | 39 45 1 0 0 0 0 | + | 39 45 1 0 0 0 0 |
| − | 40 46 1 0 0 0 0 | + | 40 46 1 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 31 18 1 0 0 0 0 | + | 31 18 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 33 47 1 0 0 0 0 | + | 33 47 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 47 48 | + | M SAL 1 2 47 48 |
| − | M SBL 1 1 52 | + | M SBL 1 1 52 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 52 0.6799 -0.6440 | + | M SBV 1 52 0.6799 -0.6440 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAGGI0001 | + | ID FL3FAGGI0001 |
| − | FORMULA C32H40O16 | + | FORMULA C32H40O16 |
| − | EXACTMASS 680.2316352319999 | + | EXACTMASS 680.2316352319999 |
| − | AVERAGEMASS 680.6504 | + | AVERAGEMASS 680.6504 |
| − | SMILES O=C(C=4CCC(C)C)c(c(OC4c(c5)cc(O)c(O)c(O)5)1)c(OC(C2OC(O3)C(C(C(C(C)3)O)O)O)OC(CO)C(O)C2O)cc(O)c1 | + | SMILES O=C(C=4CCC(C)C)c(c(OC4c(c5)cc(O)c(O)c(O)5)1)c(OC(C2OC(O3)C(C(C(C(C)3)O)O)O)OC(CO)C(O)C2O)cc(O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
48 52 0 0 0 0 0 0 0 0999 V2000
-1.1905 1.6266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1905 0.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4897 0.3484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2114 0.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2114 1.5625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4897 1.9671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9124 0.3484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6133 0.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6133 1.5625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9124 1.9671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9124 -0.2827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3141 1.9671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0286 1.5545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7430 1.9671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7430 2.7920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0286 3.2045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3141 2.7920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4897 -0.4594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4573 3.2045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4573 1.5547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9074 2.0405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0286 4.0294 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3130 0.3492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3130 -0.4585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0125 -0.8624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7119 -0.4585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0125 -1.6702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7832 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1421 -1.2755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2190 -0.9165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3282 -0.9069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9756 -0.2594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9185 -0.5981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4573 -0.6795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0988 -1.1399 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4000 -1.4552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0771 -2.3813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5592 -3.2161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6323 -2.9512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6999 -3.2161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2179 -2.3813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1447 -2.6461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9709 -2.5194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1602 -3.0550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2182 -3.6783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6999 -4.0294 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5984 0.0460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7101 -0.4669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
15 19 1 0 0 0 0
14 20 1 0 0 0 0
21 1 1 0 0 0 0
16 22 1 0 0 0 0
8 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
25 27 1 0 0 0 0
28 29 1 1 0 0 0
29 30 1 1 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 28 1 0 0 0 0
28 34 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 1 0 0 0
39 40 1 1 0 0 0
41 40 1 1 0 0 0
41 42 1 0 0 0 0
42 37 1 0 0 0 0
37 43 1 0 0 0 0
38 44 1 0 0 0 0
39 45 1 0 0 0 0
40 46 1 0 0 0 0
41 36 1 0 0 0 0
31 18 1 0 0 0 0
47 48 1 0 0 0 0
33 47 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 47 48
M SBL 1 1 52
M SMT 1 ^CH2OH
M SBV 1 52 0.6799 -0.6440
S SKP 5
ID FL3FAGGI0001
FORMULA C32H40O16
EXACTMASS 680.2316352319999
AVERAGEMASS 680.6504
SMILES O=C(C=4CCC(C)C)c(c(OC4c(c5)cc(O)c(O)c(O)5)1)c(OC(C2OC(O3)C(C(C(C(C)3)O)O)O)OC(CO)C(O)C2O)cc(O)c1
M END