Mol:FL3FAICS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2046 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2046 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2046 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2046 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6483 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6483 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0920 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0920 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0920 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0920 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6483 -0.6810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6483 -0.6810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4643 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4643 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0206 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0206 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0206 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0206 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4643 -0.6810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4643 -0.6810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4643 -2.4665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4643 -2.4665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7607 -0.6811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7607 -0.6811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4236 1.7972 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -0.4236 1.7972 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -1.0291 1.3385 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.0291 1.3385 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.7722 0.6778 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.7722 0.6778 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.7654 0.0404 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.7654 0.0404 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.3020 0.5036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3020 0.5036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6071 1.0815 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.6071 1.0815 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.8916 2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8916 2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0640 2.0536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0640 2.0536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4077 0.2993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4077 0.2993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6483 -2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6483 -2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7004 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7004 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2866 -0.9164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2866 -0.9164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8728 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8728 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8728 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8728 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2866 0.4374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2866 0.4374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7004 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7004 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7388 0.5989 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7388 0.5989 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3021 -1.7064 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.3021 -1.7064 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.9309 -2.1964 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.9309 -2.1964 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.3964 -1.9885 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.3964 -1.9885 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.8807 -1.9829 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.8807 -1.9829 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.2555 -1.6081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2555 -1.6081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7231 -1.8549 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.7231 -1.8549 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.8930 -1.6547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8930 -1.6547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2814 -2.6576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2814 -2.6576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0902 -2.5026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0902 -2.5026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0318 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0318 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6827 1.0012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6827 1.0012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4156 -1.3125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4156 -1.3125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9990 -0.7292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9990 -0.7292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7388 -1.0780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7388 -1.0780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6048 -1.5781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6048 -1.5781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5796 1.0190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5796 1.0190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0295 1.9121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0295 1.9121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 9 23 1 0 0 0 0 | + | 9 23 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 2 1 0 0 0 0 | + | 33 2 1 0 0 0 0 |
| − | 18 39 1 0 0 0 0 | + | 18 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 25 43 1 0 0 0 0 | + | 25 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 27 45 1 0 0 0 0 | + | 27 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 41 42 | + | M SAL 4 2 41 42 |
| − | M SBL 4 1 45 | + | M SBL 4 1 45 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 45 -3.1215 -1.0707 | + | M SVB 4 45 -3.1215 -1.0707 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 39 40 | + | M SAL 3 2 39 40 |
| − | M SBL 3 1 43 | + | M SBL 3 1 43 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 43 -0.0318 1.4137 | + | M SVB 3 43 -0.0318 1.4137 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 45 46 | + | M SAL 2 2 45 46 |
| − | M SBL 2 1 49 | + | M SBL 2 1 49 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 49 2.5796 1.019 | + | M SVB 2 49 2.5796 1.019 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 47 3.1015 -0.7842 | + | M SVB 1 47 3.1015 -0.7842 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAICS0005 | + | ID FL3FAICS0005 |
| − | KNApSAcK_ID C00006295 | + | KNApSAcK_ID C00006295 |
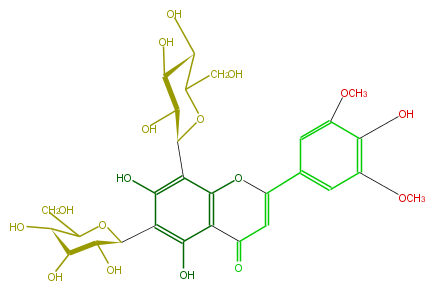
| − | NAME 6,8-Di-C-glucopyranosyltricin | + | NAME 6,8-Di-C-glucopyranosyltricin |
| − | CAS_RN 79504-25-9 | + | CAS_RN 79504-25-9 |
| − | FORMULA C29H34O17 | + | FORMULA C29H34O17 |
| − | EXACTMASS 654.179599662 | + | EXACTMASS 654.179599662 |
| − | AVERAGEMASS 654.57006 | + | AVERAGEMASS 654.57006 |
| − | SMILES [C@H](O)([C@@H]1c(c24)c(c([C@H](O5)[C@@H](O)[C@H]([C@H](C5CO)O)O)c(c2C(C=C(O4)c(c3)cc(OC)c(O)c(OC)3)=O)O)O)[C@@H](O)[C@H](C(O1)CO)O | + | SMILES [C@H](O)([C@@H]1c(c24)c(c([C@H](O5)[C@@H](O)[C@H]([C@H](C5CO)O)O)c(c2C(C=C(O4)c(c3)cc(OC)c(O)c(OC)3)=O)O)O)[C@@H](O)[C@H](C(O1)CO)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-1.2046 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2046 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6483 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0920 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0920 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6483 -0.6810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4643 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0206 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0206 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4643 -0.6810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4643 -2.4665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7607 -0.6811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4236 1.7972 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.0291 1.3385 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.7722 0.6778 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.7654 0.0404 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.3020 0.5036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6071 1.0815 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.8916 2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0640 2.0536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4077 0.2993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6483 -2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7004 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2866 -0.9164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8728 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8728 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2866 0.4374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7004 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7388 0.5989 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3021 -1.7064 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.9309 -2.1964 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3964 -1.9885 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.8807 -1.9829 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.2555 -1.6081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7231 -1.8549 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.8930 -1.6547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2814 -2.6576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0902 -2.5026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0318 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6827 1.0012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4156 -1.3125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9990 -0.7292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7388 -1.0780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6048 -1.5781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5796 1.0190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0295 1.9121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
9 23 1 0 0 0 0
26 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 2 1 0 0 0 0
18 39 1 0 0 0 0
39 40 1 0 0 0 0
35 41 1 0 0 0 0
41 42 1 0 0 0 0
25 43 1 0 0 0 0
43 44 1 0 0 0 0
27 45 1 0 0 0 0
45 46 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 41 42
M SBL 4 1 45
M SMT 4 CH2OH
M SVB 4 45 -3.1215 -1.0707
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 39 40
M SBL 3 1 43
M SMT 3 CH2OH
M SVB 3 43 -0.0318 1.4137
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 45 46
M SBL 2 1 49
M SMT 2 OCH3
M SVB 2 49 2.5796 1.019
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 47
M SMT 1 OCH3
M SVB 1 47 3.1015 -0.7842
S SKP 8
ID FL3FAICS0005
KNApSAcK_ID C00006295
NAME 6,8-Di-C-glucopyranosyltricin
CAS_RN 79504-25-9
FORMULA C29H34O17
EXACTMASS 654.179599662
AVERAGEMASS 654.57006
SMILES [C@H](O)([C@@H]1c(c24)c(c([C@H](O5)[C@@H](O)[C@H]([C@H](C5CO)O)O)c(c2C(C=C(O4)c(c3)cc(OC)c(O)c(OC)3)=O)O)O)[C@@H](O)[C@H](C(O1)CO)O
M END