Mol:FL3FAIGS0009
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.1915 0.7826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1915 0.7826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1915 0.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1915 0.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2596 -0.0399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2596 -0.0399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7106 0.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7106 0.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7106 0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7106 0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2596 1.0018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2596 1.0018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1617 -0.0399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1617 -0.0399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6128 0.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6128 0.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6128 0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6128 0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1617 1.0018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1617 1.0018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3334 -0.4078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3334 -0.4078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0637 1.0017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0637 1.0017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5234 0.7363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5234 0.7363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9831 1.0017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9831 1.0017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9831 1.5326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9831 1.5326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5234 1.7980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5234 1.7980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0637 1.5326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0637 1.5326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2596 -0.5596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2596 -0.5596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6527 1.0489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6527 1.0489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9641 1.6096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9641 1.6096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2167 0.7842 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.2167 0.7842 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.7011 0.1035 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.7011 0.1035 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.9586 0.3923 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.9586 0.3923 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.2421 0.4000 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.2421 0.4000 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.7627 0.9207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7627 0.9207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5212 0.6484 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.5212 0.6484 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.7932 0.4513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7932 0.4513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5049 0.0644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5049 0.0644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5331 -0.3219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5331 -0.3219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6210 1.4899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6210 1.4899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4423 1.1802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4423 1.1802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2886 -1.2371 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.2886 -1.2371 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -1.7730 -1.9177 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.7730 -1.9177 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.0305 -1.6290 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.0305 -1.6290 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.3140 -1.6212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.3140 -1.6212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.8346 -1.1005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8346 -1.1005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5931 -1.3729 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -1.5931 -1.3729 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -2.8651 -1.5699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8651 -1.5699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5768 -1.9569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5768 -1.9569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6050 -2.3432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6050 -2.3432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8663 -0.7789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8663 -0.7789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5142 -0.8411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5142 -0.8411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2535 2.3245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2535 2.3245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1815 3.1464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1815 3.1464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8491 0.5016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8491 0.5016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7151 0.0016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7151 0.0016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 1 1 0 0 0 0 | + | 19 1 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 26 31 1 0 0 0 0 | + | 26 31 1 0 0 0 0 |
| − | 31 30 1 0 0 0 0 | + | 31 30 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 37 42 1 0 0 0 0 | + | 37 42 1 0 0 0 0 |
| − | 42 41 1 0 0 0 0 | + | 42 41 1 0 0 0 0 |
| − | 35 18 1 0 0 0 0 | + | 35 18 1 0 0 0 0 |
| − | 16 43 1 0 0 0 0 | + | 16 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 14 45 1 0 0 0 0 | + | 14 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 42 41 | + | M SAL 4 2 42 41 |
| − | M SBL 4 1 44 | + | M SBL 4 1 44 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 44 -2.5142 -0.8411 | + | M SVB 4 44 -2.5142 -0.8411 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 31 30 | + | M SAL 3 2 31 30 |
| − | M SBL 3 1 32 | + | M SBL 3 1 32 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 32 -3.4423 1.1802 | + | M SVB 3 32 -3.4423 1.1802 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 45 46 | + | M SAL 2 2 45 46 |
| − | M SBL 2 1 49 | + | M SBL 2 1 49 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 49 3.0855 0.7955 | + | M SVB 2 49 3.0855 0.7955 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 47 2.7532 2.3432 | + | M SVB 1 47 2.7532 2.3432 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAIGS0009 | + | ID FL3FAIGS0009 |
| − | KNApSAcK_ID C00004467 | + | KNApSAcK_ID C00004467 |
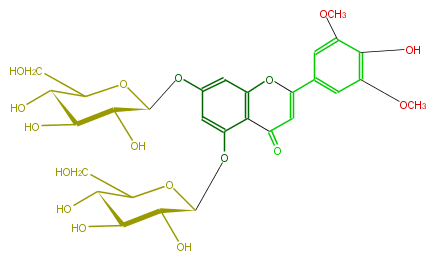
| − | NAME Tricin 5,7-diglucoside | + | NAME Tricin 5,7-diglucoside |
| − | CAS_RN 74336-88-2 | + | CAS_RN 74336-88-2 |
| − | FORMULA C29H34O17 | + | FORMULA C29H34O17 |
| − | EXACTMASS 654.179599662 | + | EXACTMASS 654.179599662 |
| − | AVERAGEMASS 654.57006 | + | AVERAGEMASS 654.57006 |
| − | SMILES [C@H](O)([C@H](Oc(c42)cc(O[C@H](O5)[C@H]([C@H]([C@@H](O)C5CO)O)O)cc2OC(=CC4=O)c(c3)cc(OC)c(c3OC)O)1)[C@H]([C@H](C(O1)CO)O)O | + | SMILES [C@H](O)([C@H](Oc(c42)cc(O[C@H](O5)[C@H]([C@H]([C@@H](O)C5CO)O)O)cc2OC(=CC4=O)c(c3)cc(OC)c(c3OC)O)1)[C@H]([C@H](C(O1)CO)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-0.1915 0.7826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1915 0.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2596 -0.0399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7106 0.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7106 0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2596 1.0018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1617 -0.0399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6128 0.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6128 0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1617 1.0018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3334 -0.4078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0637 1.0017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5234 0.7363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9831 1.0017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9831 1.5326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5234 1.7980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0637 1.5326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2596 -0.5596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6527 1.0489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9641 1.6096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2167 0.7842 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.7011 0.1035 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.9586 0.3923 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.2421 0.4000 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.7627 0.9207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5212 0.6484 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.7932 0.4513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5049 0.0644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5331 -0.3219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6210 1.4899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4423 1.1802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2886 -1.2371 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.7730 -1.9177 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.0305 -1.6290 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.3140 -1.6212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.8346 -1.1005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5931 -1.3729 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-2.8651 -1.5699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5768 -1.9569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6050 -2.3432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8663 -0.7789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5142 -0.8411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2535 2.3245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1815 3.1464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8491 0.5016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7151 0.0016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 1 1 0 0 0 0
20 15 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
26 31 1 0 0 0 0
31 30 1 0 0 0 0
24 19 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
37 42 1 0 0 0 0
42 41 1 0 0 0 0
35 18 1 0 0 0 0
16 43 1 0 0 0 0
43 44 1 0 0 0 0
14 45 1 0 0 0 0
45 46 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 42 41
M SBL 4 1 44
M SMT 4 CH2OH
M SVB 4 44 -2.5142 -0.8411
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 31 30
M SBL 3 1 32
M SMT 3 CH2OH
M SVB 3 32 -3.4423 1.1802
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 45 46
M SBL 2 1 49
M SMT 2 OCH3
M SVB 2 49 3.0855 0.7955
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 47
M SMT 1 OCH3
M SVB 1 47 2.7532 2.3432
S SKP 8
ID FL3FAIGS0009
KNApSAcK_ID C00004467
NAME Tricin 5,7-diglucoside
CAS_RN 74336-88-2
FORMULA C29H34O17
EXACTMASS 654.179599662
AVERAGEMASS 654.57006
SMILES [C@H](O)([C@H](Oc(c42)cc(O[C@H](O5)[C@H]([C@H]([C@@H](O)C5CO)O)O)cc2OC(=CC4=O)c(c3)cc(OC)c(c3OC)O)1)[C@H]([C@H](C(O1)CO)O)O
M END