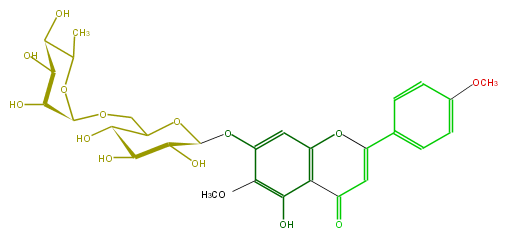
Mol:FL3FEAGS0024
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.3732 -0.4042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3732 -0.4042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3732 -1.2134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3732 -1.2134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3277 -1.6180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3277 -1.6180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0285 -1.2134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0285 -1.2134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0285 -0.4042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0285 -0.4042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3277 0.0006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3277 0.0006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7294 -1.6180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7294 -1.6180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4302 -1.2134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4302 -1.2134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4302 -0.4042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4302 -0.4042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7294 0.0006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7294 0.0006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7294 -2.3148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7294 -2.3148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1308 0.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1308 0.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8452 -0.4121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8452 -0.4121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5595 0.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5595 0.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5595 0.8252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5595 0.8252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8452 1.2376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8452 1.2376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1308 0.8252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1308 0.8252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3279 -2.3153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3279 -2.3153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0694 -0.0074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0694 -0.0074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9944 0.1419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9944 0.1419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4066 -0.6339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4066 -0.6339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5603 -0.3047 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5603 -0.3047 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7436 -0.2959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7436 -0.2959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3370 0.2976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3370 0.2976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0775 -0.0932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0775 -0.0932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4821 0.3754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4821 0.3754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6210 -0.1436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6210 -0.1436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0533 -0.6387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0533 -0.6387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8884 -0.7696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8884 -0.7696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6790 0.7074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6790 0.7074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9629 0.2940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9629 0.2940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1901 1.0890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1901 1.0890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9629 1.8889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9629 1.8889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6790 2.3025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6790 2.3025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4518 1.5073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4518 1.5073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2055 0.4690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2055 0.4690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3201 3.0050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3201 3.0050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9065 2.5375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9065 2.5375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1358 1.9673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1358 1.9673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.3018 0.7076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.3018 0.7076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0520 -1.5449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0520 -1.5449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5834 -3.0050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5834 -3.0050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1917 1.2661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1917 1.2661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.3018 0.6251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.3018 0.6251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 3 1 0 0 0 0 | + | 18 3 1 0 0 0 0 |
| − | 19 1 1 0 0 0 0 | + | 19 1 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 20 27 1 0 0 0 0 | + | 20 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 30 1 1 0 0 0 | + | 35 30 1 1 0 0 0 |
| − | 31 36 1 0 0 0 0 | + | 31 36 1 0 0 0 0 |
| − | 36 26 1 0 0 0 0 | + | 36 26 1 0 0 0 0 |
| − | 34 37 1 0 0 0 0 | + | 34 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 30 40 1 0 0 0 0 | + | 30 40 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 2 41 1 0 0 0 0 | + | 2 41 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 15 43 1 0 0 0 0 | + | 15 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^ OCH3 | + | M SMT 1 ^ OCH3 |
| − | M SBV 1 46 0.6787 0.3314 | + | M SBV 1 46 0.6787 0.3314 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 48 -0.6322 -0.4409 | + | M SBV 2 48 -0.6322 -0.4409 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FEAGS0024 | + | ID FL3FEAGS0024 |
| − | FORMULA C29H34O15 | + | FORMULA C29H34O15 |
| − | EXACTMASS 622.189770418 | + | EXACTMASS 622.189770418 |
| − | AVERAGEMASS 622.5712599999999 | + | AVERAGEMASS 622.5712599999999 |
| − | SMILES c(c5)(ccc(OC)c5)C(=C1)Oc(c2)c(c(O)c(c2OC(C(O)3)OC(COC(O4)C(C(C(O)C(C)4)O)O)C(O)C(O)3)OC)C1=O | + | SMILES c(c5)(ccc(OC)c5)C(=C1)Oc(c2)c(c(O)c(c2OC(C(O)3)OC(COC(O4)C(C(C(O)C(C)4)O)O)C(O)C(O)3)OC)C1=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-0.3732 -0.4042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3732 -1.2134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3277 -1.6180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0285 -1.2134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0285 -0.4042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3277 0.0006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7294 -1.6180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4302 -1.2134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4302 -0.4042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7294 0.0006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7294 -2.3148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1308 0.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8452 -0.4121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5595 0.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5595 0.8252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8452 1.2376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1308 0.8252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3279 -2.3153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0694 -0.0074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9944 0.1419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4066 -0.6339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5603 -0.3047 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7436 -0.2959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3370 0.2976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0775 -0.0932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4821 0.3754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6210 -0.1436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0533 -0.6387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8884 -0.7696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.6790 0.7074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9629 0.2940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1901 1.0890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9629 1.8889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6790 2.3025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4518 1.5073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2055 0.4690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3201 3.0050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9065 2.5375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.1358 1.9673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.3018 0.7076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0520 -1.5449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5834 -3.0050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1917 1.2661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.3018 0.6251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 3 1 0 0 0 0
19 1 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
25 26 1 0 0 0 0
20 27 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 30 1 1 0 0 0
31 36 1 0 0 0 0
36 26 1 0 0 0 0
34 37 1 0 0 0 0
33 38 1 0 0 0 0
35 39 1 0 0 0 0
30 40 1 0 0 0 0
23 19 1 0 0 0 0
41 42 1 0 0 0 0
2 41 1 0 0 0 0
43 44 1 0 0 0 0
15 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 ^ OCH3
M SBV 1 46 0.6787 0.3314
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 48
M SMT 2 OCH3
M SBV 2 48 -0.6322 -0.4409
S SKP 5
ID FL3FEAGS0024
FORMULA C29H34O15
EXACTMASS 622.189770418
AVERAGEMASS 622.5712599999999
SMILES c(c5)(ccc(OC)c5)C(=C1)Oc(c2)c(c(O)c(c2OC(C(O)3)OC(COC(O4)C(C(C(O)C(C)4)O)O)C(O)C(O)3)OC)C1=O
M END