Mol:FL4DAAGI0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 38 41 0 0 0 0 0 0 0 0999 V2000 | + | 38 41 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.6372 0.0741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6372 0.0741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1163 -0.2266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1163 -0.2266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4045 0.0741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4045 0.0741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4045 0.6756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4045 0.6756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1163 0.9763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1163 0.9763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6372 0.6756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6372 0.6756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9254 -0.2266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9254 -0.2266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4463 0.0741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4463 0.0741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4463 0.6756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4463 0.6756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9254 0.9763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9254 0.9763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9254 -0.7506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9254 -0.7506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0260 0.9750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0260 0.9750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5547 0.6697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5547 0.6697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0835 0.9750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0835 0.9750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0835 1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0835 1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5547 1.8908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5547 1.8908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0260 1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0260 1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1704 0.9834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1704 0.9834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9145 -0.5026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9145 -0.5026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6122 1.8908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6122 1.8908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2129 0.8183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2129 0.8183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8688 0.3641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8688 0.3641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3733 0.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3733 0.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8952 0.5619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8952 0.5619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2426 0.9094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2426 0.9094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7488 0.7277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7488 0.7277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6122 0.7113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6122 0.7113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4053 0.3379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4053 0.3379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0894 0.0801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0894 0.0801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1163 -0.8275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1163 -0.8275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1576 -0.2263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1576 -0.2263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1576 -1.0595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1576 -1.0595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6376 -1.3366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6376 -1.3366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6376 -1.8908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6376 -1.8908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2492 -1.6897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2492 -1.6897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2686 -0.9724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2686 -0.9724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5918 1.1027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5918 1.1027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3887 1.3163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3887 1.3163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 3 7 1 0 0 0 0 | + | 3 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 6 18 1 0 0 0 0 | + | 6 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 2 30 1 0 0 0 0 | + | 2 30 1 0 0 0 0 |
| − | 1 31 1 0 0 0 0 | + | 1 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 33 36 1 0 0 0 0 | + | 33 36 1 0 0 0 0 |
| − | 26 37 1 0 0 0 0 | + | 26 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 37 38 | + | M SAL 1 2 37 38 |
| − | M SBL 1 1 40 | + | M SBL 1 1 40 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 40 -6.7312 4.6781 | + | M SBV 1 40 -6.7312 4.6781 |
| − | S SKP 8 | + | S SKP 8 |
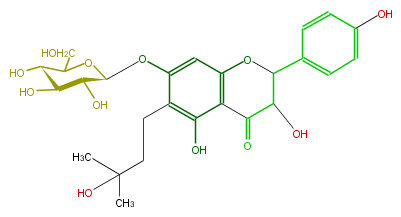
| − | ID FL4DAAGI0002 | + | ID FL4DAAGI0002 |
| − | KNApSAcK_ID C00008728 | + | KNApSAcK_ID C00008728 |
| − | NAME Phellavin | + | NAME Phellavin |
| − | CAS_RN 32507-67-8 | + | CAS_RN 32507-67-8 |
| − | FORMULA C26H32O12 | + | FORMULA C26H32O12 |
| − | EXACTMASS 536.189376488 | + | EXACTMASS 536.189376488 |
| − | AVERAGEMASS 536.5250799999999 | + | AVERAGEMASS 536.5250799999999 |
| − | SMILES Oc(c4)ccc(c4)C(O3)C(C(=O)c(c23)c(c(c(c2)OC(C1O)OC(CO)C(O)C1O)CCC(C)(C)O)O)O | + | SMILES Oc(c4)ccc(c4)C(O3)C(C(=O)c(c23)c(c(c(c2)OC(C1O)OC(CO)C(O)C1O)CCC(C)(C)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
38 41 0 0 0 0 0 0 0 0999 V2000
-0.6372 0.0741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1163 -0.2266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4045 0.0741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4045 0.6756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1163 0.9763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6372 0.6756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9254 -0.2266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4463 0.0741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4463 0.6756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9254 0.9763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9254 -0.7506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0260 0.9750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5547 0.6697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0835 0.9750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0835 1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5547 1.8908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0260 1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1704 0.9834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9145 -0.5026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6122 1.8908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2129 0.8183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8688 0.3641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3733 0.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8952 0.5619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2426 0.9094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7488 0.7277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6122 0.7113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4053 0.3379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0894 0.0801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1163 -0.8275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1576 -0.2263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1576 -1.0595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6376 -1.3366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6376 -1.8908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2492 -1.6897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2686 -0.9724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5918 1.1027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3887 1.3163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
3 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 4 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
6 18 1 0 0 0 0
8 19 1 0 0 0 0
15 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 18 1 0 0 0 0
2 30 1 0 0 0 0
1 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
33 35 1 0 0 0 0
33 36 1 0 0 0 0
26 37 1 0 0 0 0
37 38 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 37 38
M SBL 1 1 40
M SMT 1 ^CH2OH
M SBV 1 40 -6.7312 4.6781
S SKP 8
ID FL4DAAGI0002
KNApSAcK_ID C00008728
NAME Phellavin
CAS_RN 32507-67-8
FORMULA C26H32O12
EXACTMASS 536.189376488
AVERAGEMASS 536.5250799999999
SMILES Oc(c4)ccc(c4)C(O3)C(C(=O)c(c23)c(c(c(c2)OC(C1O)OC(CO)C(O)C1O)CCC(C)(C)O)O)O
M END