Mol:FL5FAAGA0007
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.9140 0.9040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9140 0.9040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9140 0.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9140 0.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2130 -0.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2130 -0.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5120 0.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5120 0.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5120 0.9040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5120 0.9040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2130 1.3086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2130 1.3086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8110 -0.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8110 -0.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1100 0.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1100 0.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1100 0.9040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1100 0.9040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8110 1.3086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8110 1.3086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8110 -0.9413 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8110 -0.9413 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4093 1.3085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4093 1.3085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3052 0.8960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3052 0.8960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0197 1.3085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0197 1.3085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0197 2.1335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0197 2.1335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3052 2.5460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3052 2.5460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4093 2.1335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4093 2.1335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2130 -1.1194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2130 -1.1194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6148 1.3085 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6148 1.3085 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7339 2.5459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7339 2.5459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5551 -0.6237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5551 -0.6237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4027 -0.1964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4027 -0.1964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3029 -0.0376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3029 -0.0376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5455 0.4742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5455 0.4742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1696 1.3130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1696 1.3130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2693 1.1544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2693 1.1544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0266 0.6424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0266 0.6424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7640 1.2453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7640 1.2453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0462 -0.6470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0462 -0.6470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6148 -0.7065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6148 -0.7065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2483 0.4777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2483 0.4777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8938 -0.2393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8938 -0.2393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0478 0.2492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0478 0.2492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3162 -0.6902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3162 -0.6902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0478 -1.6353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0478 -1.6353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8938 -2.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8938 -2.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6254 -1.1844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6254 -1.1844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8460 0.5140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8460 0.5140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2954 -1.5712 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2954 -1.5712 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5138 -2.1046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5138 -2.1046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1531 -2.5460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1531 -2.5460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1489 -2.2769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1489 -2.2769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 21 33 1 0 0 0 0 | + | 21 33 1 0 0 0 0 |
| − | 26 38 1 0 0 0 0 | + | 26 38 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 46 -0.1053 0.9107 | + | M SBV 1 46 -0.1053 0.9107 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGA0007 | + | ID FL5FAAGA0007 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
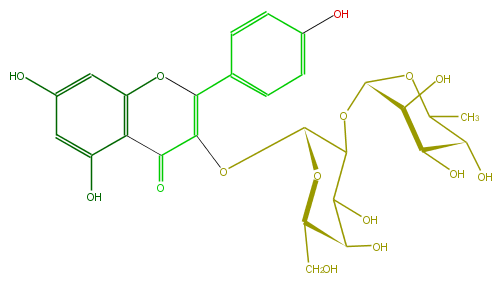
| − | SMILES C(C5O)(OC(C)C(C5O)O)OC(C(OC(C4=O)=C(Oc(c43)cc(O)cc3O)c(c2)ccc(c2)O)1)C(O)C(C(CO)O1)O | + | SMILES C(C5O)(OC(C)C(C5O)O)OC(C(OC(C4=O)=C(Oc(c43)cc(O)cc3O)c(c2)ccc(c2)O)1)C(O)C(C(CO)O1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-3.9140 0.9040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9140 0.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2130 -0.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5120 0.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5120 0.9040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2130 1.3086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8110 -0.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1100 0.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1100 0.9040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8110 1.3086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8110 -0.9413 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4093 1.3085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3052 0.8960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0197 1.3085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0197 2.1335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3052 2.5460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4093 2.1335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2130 -1.1194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6148 1.3085 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7339 2.5459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5551 -0.6237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4027 -0.1964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3029 -0.0376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5455 0.4742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1696 1.3130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2693 1.1544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0266 0.6424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7640 1.2453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0462 -0.6470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6148 -0.7065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2483 0.4777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8938 -0.2393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0478 0.2492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3162 -0.6902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0478 -1.6353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8938 -2.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6254 -1.1844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8460 0.5140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2954 -1.5712 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5138 -2.1046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1531 -2.5460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1489 -2.2769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
21 8 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
27 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
37 39 1 0 0 0 0
36 40 1 0 0 0 0
21 33 1 0 0 0 0
26 38 1 0 0 0 0
41 42 1 0 0 0 0
35 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 CH2OH
M SBV 1 46 -0.1053 0.9107
S SKP 5
ID FL5FAAGA0007
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES C(C5O)(OC(C)C(C5O)O)OC(C(OC(C4=O)=C(Oc(c43)cc(O)cc3O)c(c2)ccc(c2)O)1)C(O)C(C(CO)O1)O
M END