Mol:FL5FAAGA0009
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0956 0.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0956 0.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0956 0.1563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0956 0.1563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5393 -0.1649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5393 -0.1649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9830 0.1563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9830 0.1563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9830 0.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9830 0.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5393 1.1199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5393 1.1199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4267 -0.1649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4267 -0.1649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1296 0.1563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1296 0.1563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1296 0.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1296 0.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4267 1.1199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4267 1.1199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4267 -0.6657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4267 -0.6657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6857 1.1197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6857 1.1197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2527 0.7924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2527 0.7924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8196 1.1197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8196 1.1197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8196 1.7744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8196 1.7744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2527 2.1018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2527 2.1018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6857 1.7744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6857 1.7744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5393 -0.8070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5393 -0.8070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6517 1.1197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6517 1.1197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3864 2.1017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3864 2.1017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7983 -0.3527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7983 -0.3527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8437 0.1387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8437 0.1387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4561 -0.5328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4561 -0.5328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2016 -0.3197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2016 -0.3197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9515 -0.5328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9515 -0.5328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3393 0.1387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3393 0.1387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5938 -0.0743 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5938 -0.0743 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1238 -0.0542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1238 -0.0542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9007 0.4573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9007 0.4573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3393 1.0476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3393 1.0476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0963 -0.9545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0963 -0.9545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6838 -1.6557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6838 -1.6557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6319 -1.3876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6319 -1.3876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9154 -1.3799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9154 -1.3799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4360 -0.8591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4360 -0.8591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0856 -1.2020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0856 -1.2020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0963 -0.3137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0963 -0.3137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1782 -1.7155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1782 -1.7155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2064 -2.1018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2064 -2.1018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8227 -0.8647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8227 -0.8647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6196 -0.6511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6196 -0.6511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4637 -0.5060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4637 -0.5060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1782 -0.9185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1782 -0.9185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 18 1 0 0 0 0 | + | 34 18 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 42 1 0 0 0 0 | + | 25 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 44 -7.8511 4.1266 | + | M SBV 1 44 -7.8511 4.1266 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 46 -7.6018 3.8160 | + | M SBV 2 46 -7.6018 3.8160 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGA0009 | + | ID FL5FAAGA0009 |
| − | KNApSAcK_ID C00005173 | + | KNApSAcK_ID C00005173 |
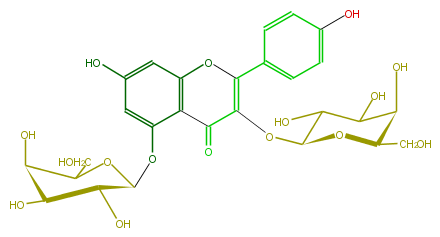
| − | NAME Kaempferol 3,5-digalactoside | + | NAME Kaempferol 3,5-digalactoside |
| − | CAS_RN 91377-10-5 | + | CAS_RN 91377-10-5 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES O(C=1c(c5)ccc(c5)O)c(c4)c(c(cc(O)4)OC(O3)C(C(O)C(C3CO)O)O)C(=O)C1OC(C(O)2)OC(C(C(O)2)O)CO | + | SMILES O(C=1c(c5)ccc(c5)O)c(c4)c(c(cc(O)4)OC(O3)C(C(O)C(C3CO)O)O)C(=O)C1OC(C(O)2)OC(C(C(O)2)O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-2.0956 0.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0956 0.1563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5393 -0.1649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9830 0.1563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9830 0.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5393 1.1199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4267 -0.1649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1296 0.1563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1296 0.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4267 1.1199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4267 -0.6657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6857 1.1197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2527 0.7924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8196 1.1197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8196 1.7744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2527 2.1018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6857 1.7744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5393 -0.8070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6517 1.1197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3864 2.1017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7983 -0.3527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8437 0.1387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4561 -0.5328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2016 -0.3197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9515 -0.5328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3393 0.1387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5938 -0.0743 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1238 -0.0542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9007 0.4573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3393 1.0476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0963 -0.9545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6838 -1.6557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6319 -1.3876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9154 -1.3799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4360 -0.8591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0856 -1.2020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0963 -0.3137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1782 -1.7155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2064 -2.1018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8227 -0.8647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6196 -0.6511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4637 -0.5060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1782 -0.9185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
8 21 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 18 1 0 0 0 0
36 40 1 0 0 0 0
40 41 1 0 0 0 0
25 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 ^CH2OH
M SBV 1 44 -7.8511 4.1266
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 46
M SMT 2 CH2OH
M SBV 2 46 -7.6018 3.8160
S SKP 8
ID FL5FAAGA0009
KNApSAcK_ID C00005173
NAME Kaempferol 3,5-digalactoside
CAS_RN 91377-10-5
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES O(C=1c(c5)ccc(c5)O)c(c4)c(c(cc(O)4)OC(O3)C(C(O)C(C3CO)O)O)C(=O)C1OC(C(O)2)OC(C(C(O)2)O)CO
M END