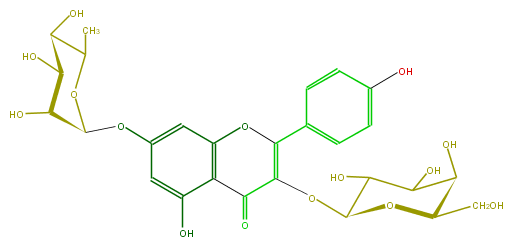
Mol:FL5FAAGA0011
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.6714 -0.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6714 -0.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6714 -1.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6714 -1.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9702 -1.6821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9702 -1.6821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2691 -1.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2691 -1.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2691 -0.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2691 -0.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9702 -0.0630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9702 -0.0630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5680 -1.6821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5680 -1.6821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1331 -1.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1331 -1.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1331 -0.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1331 -0.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5680 -0.0630 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5680 -0.0630 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5680 -2.3134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5680 -2.3134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8340 -0.0632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8340 -0.0632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5484 -0.4758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5484 -0.4758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2630 -0.0632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2630 -0.0632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2630 0.7620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2630 0.7620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5484 1.1745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5484 1.1745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8340 0.7620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8340 0.7620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9702 -2.4914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9702 -2.4914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3722 -0.0632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3722 -0.0632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9774 1.1743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9774 1.1743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9566 -1.6935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9566 -1.6935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9577 0.2227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9577 0.2227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1659 -0.2344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1659 -0.2344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4172 0.6447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4172 0.6447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1659 1.5290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1659 1.5290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9577 1.9863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9577 1.9863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7066 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7066 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4526 2.4914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4526 2.4914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1659 2.0996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1659 2.0996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3555 1.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3555 1.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6458 0.2071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6458 0.2071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9825 -0.4847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9825 -0.4847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2393 -1.2537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2393 -1.2537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7285 -2.1385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7285 -2.1385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7108 -1.8577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7108 -1.8577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6991 -2.1385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6991 -2.1385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2101 -1.2537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2101 -1.2537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2277 -1.5344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2277 -1.5344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6010 -1.2109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6010 -1.2109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6089 -1.0648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6089 -1.0648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6415 -1.8627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6415 -1.8627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6458 -2.1319 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6458 -2.1319 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 27 30 1 0 0 0 0 | + | 27 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 21 34 1 0 0 0 0 | + | 21 34 1 0 0 0 0 |
| − | 38 40 1 0 0 0 0 | + | 38 40 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 36 41 1 0 0 0 0 | + | 36 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 46 -0.9424 -0.2758 | + | M SBV 1 46 -0.9424 -0.2758 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGA0011 | + | ID FL5FAAGA0011 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES c(c25)(O)cc(cc2OC(=C(C5=O)OC(C(O)4)OC(C(C(O)4)O)CO)c(c3)ccc(c3)O)OC(C(O)1)OC(C)C(O)C1O | + | SMILES c(c25)(O)cc(cc2OC(=C(C5=O)OC(C(O)4)OC(C(C(O)4)O)CO)c(c3)ccc(c3)O)OC(C(O)1)OC(C)C(O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-2.6714 -0.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6714 -1.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9702 -1.6821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2691 -1.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2691 -0.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9702 -0.0630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5680 -1.6821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1331 -1.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1331 -0.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5680 -0.0630 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5680 -2.3134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8340 -0.0632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5484 -0.4758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2630 -0.0632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2630 0.7620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5484 1.1745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8340 0.7620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9702 -2.4914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3722 -0.0632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9774 1.1743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9566 -1.6935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9577 0.2227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1659 -0.2344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4172 0.6447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1659 1.5290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9577 1.9863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7066 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4526 2.4914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1659 2.0996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3555 1.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.6458 0.2071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9825 -0.4847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2393 -1.2537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7285 -2.1385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7108 -1.8577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6991 -2.1385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2101 -1.2537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2277 -1.5344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6010 -1.2109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6089 -1.0648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6415 -1.8627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6458 -2.1319 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
27 30 1 0 0 0 0
22 31 1 0 0 0 0
23 19 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
37 32 1 0 0 0 0
33 39 1 0 0 0 0
21 34 1 0 0 0 0
38 40 1 0 0 0 0
41 42 1 0 0 0 0
36 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 CH2OH
M SBV 1 46 -0.9424 -0.2758
S SKP 5
ID FL5FAAGA0011
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES c(c25)(O)cc(cc2OC(=C(C5=O)OC(C(O)4)OC(C(C(O)4)O)CO)c(c3)ccc(c3)O)OC(C(O)1)OC(C)C(O)C1O
M END