Mol:FL5FAAGL0007
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.3446 1.6628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3446 1.6628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3446 1.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3446 1.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7883 0.6993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7883 0.6993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2320 1.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2320 1.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2320 1.6628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2320 1.6628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7883 1.9840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7883 1.9840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6757 0.6993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6757 0.6993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1194 1.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1194 1.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1194 1.6628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1194 1.6628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6757 1.9840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6757 1.9840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6757 0.1985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6757 0.1985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5633 1.9839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5633 1.9839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0037 1.6566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0037 1.6566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5707 1.9839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5707 1.9839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5707 2.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5707 2.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0037 2.9659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0037 2.9659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5633 2.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5633 2.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7883 0.0572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7883 0.0572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9007 1.9839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9007 1.9839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1375 2.9658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1375 2.9658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6790 -1.5966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6790 -1.5966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0508 -1.9593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0508 -1.9593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2501 -1.2618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2501 -1.2618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0508 -0.5601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0508 -0.5601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6790 -0.1973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6790 -0.1973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4797 -0.8948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4797 -0.8948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6790 0.4506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6790 0.4506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2126 0.0439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2126 0.0439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2119 -1.5285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2119 -1.5285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8010 0.4872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8010 0.4872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3167 -0.1934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3167 -0.1934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0592 0.0953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0592 0.0953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7756 0.1031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7756 0.1031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2550 0.6238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2550 0.6238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6054 0.2810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6054 0.2810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5129 -0.2325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5129 -0.2325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4846 -0.6189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4846 -0.6189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9007 0.4045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9007 0.4045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3021 -2.3946 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3021 -2.3946 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6358 -2.9659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6358 -2.9659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0483 -2.2515 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0483 -2.2515 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6150 0.6097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6150 0.6097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3295 0.1972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3295 0.1972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 31 36 1 0 0 0 0 | + | 31 36 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 30 28 1 0 0 0 0 | + | 30 28 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 22 39 1 0 0 0 0 | + | 22 39 1 0 0 0 0 |
| − | 27 8 1 0 0 0 0 | + | 27 8 1 0 0 0 0 |
| − | 21 40 1 0 0 0 0 | + | 21 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 44 -7.3085 3.2016 | + | M SBV 1 44 -7.3085 3.2016 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 46 -5.9917 4.5569 | + | M SBV 2 46 -5.9917 4.5569 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGL0007 | + | ID FL5FAAGL0007 |
| − | KNApSAcK_ID C00005165 | + | KNApSAcK_ID C00005165 |
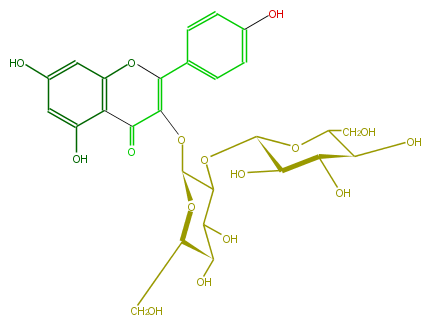
| − | NAME Kaempferol 3-sophoroside | + | NAME Kaempferol 3-sophoroside |
| − | CAS_RN 19895-95-5 | + | CAS_RN 19895-95-5 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES c(O2)(c(C(C(OC(C4OC(C5O)OC(C(C5O)O)CO)OC(CO)C(C(O)4)O)=C2c(c3)ccc(c3)O)=O)1)cc(O)cc1O | + | SMILES c(O2)(c(C(C(OC(C4OC(C5O)OC(C(C5O)O)CO)OC(CO)C(C(O)4)O)=C2c(c3)ccc(c3)O)=O)1)cc(O)cc1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-3.3446 1.6628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3446 1.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7883 0.6993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2320 1.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2320 1.6628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7883 1.9840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6757 0.6993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1194 1.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1194 1.6628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6757 1.9840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6757 0.1985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5633 1.9839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0037 1.6566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5707 1.9839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5707 2.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0037 2.9659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5633 2.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7883 0.0572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9007 1.9839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1375 2.9658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6790 -1.5966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0508 -1.9593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2501 -1.2618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0508 -0.5601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6790 -0.1973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4797 -0.8948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6790 0.4506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2126 0.0439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2119 -1.5285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8010 0.4872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3167 -0.1934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0592 0.0953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7756 0.1031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2550 0.6238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6054 0.2810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5129 -0.2325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4846 -0.6189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9007 0.4045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3021 -2.3946 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6358 -2.9659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0483 -2.2515 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6150 0.6097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3295 0.1972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
23 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
31 36 1 0 0 0 0
32 37 1 0 0 0 0
30 28 1 0 0 0 0
33 38 1 0 0 0 0
22 39 1 0 0 0 0
27 8 1 0 0 0 0
21 40 1 0 0 0 0
40 41 1 0 0 0 0
34 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 CH2OH
M SBV 1 44 -7.3085 3.2016
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 46
M SMT 2 CH2OH
M SBV 2 46 -5.9917 4.5569
S SKP 8
ID FL5FAAGL0007
KNApSAcK_ID C00005165
NAME Kaempferol 3-sophoroside
CAS_RN 19895-95-5
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES c(O2)(c(C(C(OC(C4OC(C5O)OC(C(C5O)O)CO)OC(CO)C(C(O)4)O)=C2c(c3)ccc(c3)O)=O)1)cc(O)cc1O
M END