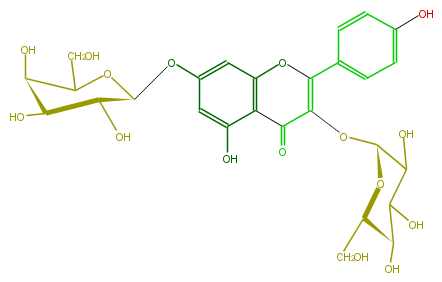
Mol:FL5FAAGL0014
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.6692 -4.8812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6692 -4.8812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3018 -4.7697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3018 -4.7697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5215 -4.1661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5215 -4.1661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1086 -3.6740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1086 -3.6740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4760 -3.7855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4760 -3.7855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2563 -4.3892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2563 -4.3892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3283 -3.0704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3283 -3.0704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9154 -2.5783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9154 -2.5783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2828 -2.6898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2828 -2.6898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0631 -3.2935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0631 -3.2935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8214 -2.9833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8214 -2.9833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8701 -2.1979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8701 -2.1979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0940 -1.5827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0940 -1.5827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6732 -1.0813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6732 -1.0813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0284 -1.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0284 -1.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1956 -1.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1956 -1.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2253 -2.3116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2253 -2.3116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1538 -4.0545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1538 -4.0545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4496 -5.4846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4496 -5.4846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3923 -0.6935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3923 -0.6935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3006 -1.8314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3006 -1.8314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7513 -0.4809 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.7513 -0.4809 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.3636 -1.1523 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.3636 -1.1523 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.1091 -0.9393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1091 -0.9393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8590 -1.1523 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.8590 -1.1523 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.2468 -0.4809 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.2468 -0.4809 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.5013 -0.6939 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.5013 -0.6939 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.0313 -0.6738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0313 -0.6738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8082 -0.1623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8082 -0.1623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7557 -0.4809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7557 -0.4809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2744 -8.3139 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 2.2744 -8.3139 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 2.8935 -7.7859 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.8935 -7.7859 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.4468 -6.7966 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.4468 -6.7966 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.3147 -6.0923 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.3147 -6.0923 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.8923 -6.6955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8923 -6.6955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3427 -7.2757 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.3427 -7.2757 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.7057 -8.4142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7057 -8.4142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0381 -8.2625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0381 -8.2625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0762 -6.2536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0762 -6.2536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6662 -7.4867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6662 -7.4867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9309 -6.8089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9309 -6.8089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6852 -1.4822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6852 -1.4822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8284 -2.4719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8284 -2.4719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 19 1 0 0 0 0 | + | 34 19 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 42 1 0 0 0 0 | + | 25 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 46 4.6852 -1.4822 | + | M SVB 2 46 4.6852 -1.4822 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 44 -3.7535 0.6345 | + | M SVB 1 44 -3.7535 0.6345 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGL0014 | + | ID FL5FAAGL0014 |
| − | KNApSAcK_ID C00005181 | + | KNApSAcK_ID C00005181 |
| − | NAME Kaempferol 3-glucoside-7-galactoside | + | NAME Kaempferol 3-glucoside-7-galactoside |
| − | CAS_RN 99883-42-8 | + | CAS_RN 99883-42-8 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES O([C@@H]([C@@H](O)5)OC(CO)[C@H](O)[C@@H]5O)c(c4)cc(c(c41)C(=O)C(O[C@@H](C(O)3)O[C@@H]([C@@H](C(O)3)O)CO)=C(c(c2)ccc(c2)O)O1)O | + | SMILES O([C@@H]([C@@H](O)5)OC(CO)[C@H](O)[C@@H]5O)c(c4)cc(c(c41)C(=O)C(O[C@@H](C(O)3)O[C@@H]([C@@H](C(O)3)O)CO)=C(c(c2)ccc(c2)O)O1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
1.6692 -4.8812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3018 -4.7697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5215 -4.1661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1086 -3.6740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4760 -3.7855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2563 -4.3892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3283 -3.0704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9154 -2.5783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2828 -2.6898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0631 -3.2935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8214 -2.9833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8701 -2.1979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0940 -1.5827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6732 -1.0813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0284 -1.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1956 -1.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2253 -2.3116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1538 -4.0545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4496 -5.4846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3923 -0.6935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3006 -1.8314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7513 -0.4809 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.3636 -1.1523 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.1091 -0.9393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8590 -1.1523 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.2468 -0.4809 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.5013 -0.6939 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.0313 -0.6738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8082 -0.1623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7557 -0.4809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2744 -8.3139 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.8935 -7.7859 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.4468 -6.7966 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.3147 -6.0923 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.8923 -6.6955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3427 -7.2757 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.7057 -8.4142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0381 -8.2625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0762 -6.2536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6662 -7.4867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9309 -6.8089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6852 -1.4822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8284 -2.4719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
8 21 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 19 1 0 0 0 0
36 40 1 0 0 0 0
40 41 1 0 0 0 0
25 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 46
M SMT 2 CH2OH
M SVB 2 46 4.6852 -1.4822
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 CH2OH
M SVB 1 44 -3.7535 0.6345
S SKP 8
ID FL5FAAGL0014
KNApSAcK_ID C00005181
NAME Kaempferol 3-glucoside-7-galactoside
CAS_RN 99883-42-8
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES O([C@@H]([C@@H](O)5)OC(CO)[C@H](O)[C@@H]5O)c(c4)cc(c(c41)C(=O)C(O[C@@H](C(O)3)O[C@@H]([C@@H](C(O)3)O)CO)=C(c(c2)ccc(c2)O)O1)O
M END