Mol:FL5FAAGL0068
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.8299 1.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8299 1.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8299 0.8916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8299 0.8916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2736 0.5704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2736 0.5704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7173 0.8916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7173 0.8916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7173 1.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7173 1.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2736 1.8551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2736 1.8551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1610 0.5704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1610 0.5704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6047 0.8916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6047 0.8916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6047 1.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6047 1.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1610 1.8551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1610 1.8551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1610 0.0696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1610 0.0696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0486 1.8550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0486 1.8550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5184 1.5277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5184 1.5277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0854 1.8550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0854 1.8550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0854 2.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0854 2.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5184 2.8371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5184 2.8371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0486 2.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0486 2.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2736 -0.0717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2736 -0.0717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8791 2.8890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8791 2.8890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3248 1.8551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3248 1.8551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9463 0.8934 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 0.9463 0.8934 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.3887 0.3094 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.3887 0.3094 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.0257 0.5571 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.0257 0.5571 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.8008 0.4821 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.8008 0.4821 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.1938 1.0106 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.1938 1.0106 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.6364 0.7164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6364 0.7164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3338 0.5398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3338 0.5398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6990 0.2758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6990 0.2758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5896 -0.0067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5896 -0.0067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8008 0.4821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8008 0.4821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5896 -0.6111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5896 -0.6111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2335 -0.9829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2335 -0.9829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9703 -0.9686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9703 -0.9686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9703 -1.6837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9703 -1.6837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3947 -2.0160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3947 -2.0160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8906 -1.7250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8906 -1.7250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3866 -2.0160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3866 -2.0160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3866 -2.5980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3866 -2.5980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8906 -2.8890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8906 -2.8890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3947 -2.5980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3947 -2.5980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1171 -2.8888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1171 -2.8888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5089 1.1356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5089 1.1356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6544 2.1249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6544 2.1249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 27 8 1 0 0 0 0 | + | 27 8 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 29 31 1 0 0 0 0 | + | 29 31 1 0 0 0 0 |
| − | 31 32 2 0 0 0 0 | + | 31 32 2 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 35 1 0 0 0 0 | + | 40 35 1 0 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 25 42 1 0 0 0 0 | + | 25 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 46 2.5322 0.9907 | + | M SVB 1 46 2.5322 0.9907 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGL0068 | + | ID FL5FAAGL0068 |
| − | KNApSAcK_ID C00005850 | + | KNApSAcK_ID C00005850 |
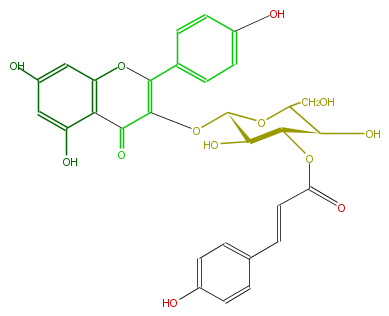
| − | NAME Kaempferol 3-(3''-p-coumarylglucoside) | + | NAME Kaempferol 3-(3''-p-coumarylglucoside) |
| − | CAS_RN 74712-68-8 | + | CAS_RN 74712-68-8 |
| − | FORMULA C30H26O13 | + | FORMULA C30H26O13 |
| − | EXACTMASS 594.137340918 | + | EXACTMASS 594.137340918 |
| − | AVERAGEMASS 594.51964 | + | AVERAGEMASS 594.51964 |
| − | SMILES O(C(CO)1)[C@H](OC(C5=O)=C(Oc(c54)cc(cc4O)O)c(c3)ccc(O)c3)[C@@H]([C@H](OC(C=Cc(c2)ccc(O)c2)=O)[C@@H]1O)O | + | SMILES O(C(CO)1)[C@H](OC(C5=O)=C(Oc(c54)cc(cc4O)O)c(c3)ccc(O)c3)[C@@H]([C@H](OC(C=Cc(c2)ccc(O)c2)=O)[C@@H]1O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-2.8299 1.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8299 0.8916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2736 0.5704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7173 0.8916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7173 1.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2736 1.8551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1610 0.5704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6047 0.8916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6047 1.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1610 1.8551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1610 0.0696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0486 1.8550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5184 1.5277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0854 1.8550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0854 2.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5184 2.8371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0486 2.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2736 -0.0717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8791 2.8890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3248 1.8551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9463 0.8934 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.3887 0.3094 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0257 0.5571 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.8008 0.4821 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.1938 1.0106 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.6364 0.7164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3338 0.5398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6990 0.2758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5896 -0.0067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8008 0.4821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5896 -0.6111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2335 -0.9829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9703 -0.9686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9703 -1.6837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3947 -2.0160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8906 -1.7250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3866 -2.0160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3866 -2.5980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8906 -2.8890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3947 -2.5980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1171 -2.8888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5089 1.1356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6544 2.1249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
27 8 1 0 0 0 0
24 30 1 0 0 0 0
29 31 1 0 0 0 0
31 32 2 0 0 0 0
31 33 1 0 0 0 0
33 34 2 0 0 0 0
34 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 35 1 0 0 0 0
38 41 1 0 0 0 0
25 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 46
M SMT 1 CH2OH
M SVB 1 46 2.5322 0.9907
S SKP 8
ID FL5FAAGL0068
KNApSAcK_ID C00005850
NAME Kaempferol 3-(3''-p-coumarylglucoside)
CAS_RN 74712-68-8
FORMULA C30H26O13
EXACTMASS 594.137340918
AVERAGEMASS 594.51964
SMILES O(C(CO)1)[C@H](OC(C5=O)=C(Oc(c54)cc(cc4O)O)c(c3)ccc(O)c3)[C@@H]([C@H](OC(C=Cc(c2)ccc(O)c2)=O)[C@@H]1O)O
M END