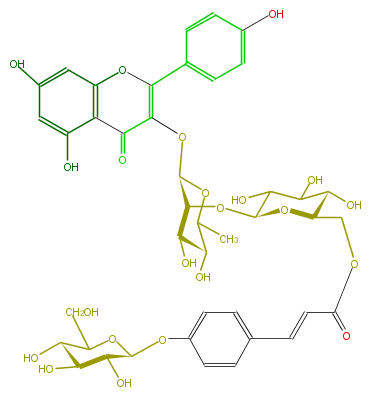
Mol:FL5FAAGS0062
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 64 70 0 0 0 0 0 0 0 0999 V2000 | + | 64 70 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.8927 2.2425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8927 2.2425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8927 1.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8927 1.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3364 1.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3364 1.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7801 1.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7801 1.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7801 2.2425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7801 2.2425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3364 2.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3364 2.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2238 1.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2238 1.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6674 1.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6674 1.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6674 2.2425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6674 2.2425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2238 2.5637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2238 2.5637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2238 0.7781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2238 0.7781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0330 2.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0330 2.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6000 2.3600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6000 2.3600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1670 2.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1670 2.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1670 3.3420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1670 3.3420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6000 3.6693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6000 3.6693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0330 3.3420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0330 3.3420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3364 0.6368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3364 0.6368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7738 3.6923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7738 3.6923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4083 2.6962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4083 2.6962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0380 1.2367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0380 1.2367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1033 -0.7794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1033 -0.7794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4321 -1.0885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4321 -1.0885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2622 -0.4941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2622 -0.4941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4321 0.1040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4321 0.1040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1033 0.4132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1033 0.4132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0665 -0.1813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0665 -0.1813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7231 -0.1813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7231 -0.1813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0342 -1.2926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0342 -1.2926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3029 -1.5706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3029 -1.5706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7908 -0.7993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7908 -0.7993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7311 0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7311 0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4303 -0.3934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4303 -0.3934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0087 -0.2281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0087 -0.2281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5907 -0.3934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5907 -0.3934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8916 0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8916 0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3131 -0.0377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3131 -0.0377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1724 -0.0221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1724 -0.0221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5513 0.3749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5513 0.3749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3257 -0.1230 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3257 -0.1230 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1423 -0.3934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1423 -0.3934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4083 -1.3050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4083 -1.3050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9942 -2.2946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9942 -2.2946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2532 -2.7432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2532 -2.7432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4385 -2.2946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4385 -2.2946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1337 -2.8225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1337 -2.8225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3246 -2.8225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3246 -2.8225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0189 -2.2930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0189 -2.2930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4074 -2.2930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4074 -2.2930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1017 -2.8225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1017 -2.8225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4074 -3.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4074 -3.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0189 -3.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0189 -3.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4303 -2.8225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4303 -2.8225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4739 -2.8961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4739 -2.8961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1027 -3.3861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1027 -3.3861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5682 -3.1782 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5682 -3.1782 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0524 -3.1727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0524 -3.1727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4272 -2.7978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4272 -2.7978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8948 -3.0446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8948 -3.0446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9877 -3.1928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9877 -3.1928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6813 -3.4142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6813 -3.4142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2619 -3.6923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2619 -3.6923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2933 -2.2604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2933 -2.2604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4964 -2.4739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4964 -2.4739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 54 55 1 1 0 0 0 | + | 54 55 1 1 0 0 0 |
| − | 55 56 1 1 0 0 0 | + | 55 56 1 1 0 0 0 |
| − | 57 56 1 1 0 0 0 | + | 57 56 1 1 0 0 0 |
| − | 57 58 1 0 0 0 0 | + | 57 58 1 0 0 0 0 |
| − | 58 59 1 0 0 0 0 | + | 58 59 1 0 0 0 0 |
| − | 59 54 1 0 0 0 0 | + | 59 54 1 0 0 0 0 |
| − | 54 60 1 0 0 0 0 | + | 54 60 1 0 0 0 0 |
| − | 55 61 1 0 0 0 0 | + | 55 61 1 0 0 0 0 |
| − | 56 62 1 0 0 0 0 | + | 56 62 1 0 0 0 0 |
| − | 57 53 1 0 0 0 0 | + | 57 53 1 0 0 0 0 |
| − | 59 63 1 0 0 0 0 | + | 59 63 1 0 0 0 0 |
| − | 63 64 1 0 0 0 0 | + | 63 64 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 63 64 | + | M SAL 1 2 63 64 |
| − | M SBL 1 1 69 | + | M SBL 1 1 69 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 69 -6.5251 7.4198 | + | M SBV 1 69 -6.5251 7.4198 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGS0062 | + | ID FL5FAAGS0062 |
| − | KNApSAcK_ID C00005885 | + | KNApSAcK_ID C00005885 |
| − | NAME Kaempferol 3-(6-[4-glucosyl-p-coumaryl]glucosyl)(1->2)-rhamnoside | + | NAME Kaempferol 3-(6-[4-glucosyl-p-coumaryl]glucosyl)(1->2)-rhamnoside |
| − | CAS_RN 142997-34-0 | + | CAS_RN 142997-34-0 |
| − | FORMULA C42H46O22 | + | FORMULA C42H46O22 |
| − | EXACTMASS 902.248073156 | + | EXACTMASS 902.248073156 |
| − | AVERAGEMASS 902.80144 | + | AVERAGEMASS 902.80144 |
| − | SMILES CC(C7O)OC(C(C7O)OC(C(O)6)OC(C(O)C6O)COC(=O)C=Cc(c5)ccc(c5)OC(O4)C(C(O)C(O)C4CO)O)OC(C(=O)1)=C(c(c3)ccc(c3)O)Oc(c2)c1c(O)cc(O)2 | + | SMILES CC(C7O)OC(C(C7O)OC(C(O)6)OC(C(O)C6O)COC(=O)C=Cc(c5)ccc(c5)OC(O4)C(C(O)C(O)C4CO)O)OC(C(=O)1)=C(c(c3)ccc(c3)O)Oc(c2)c1c(O)cc(O)2 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
64 70 0 0 0 0 0 0 0 0999 V2000
-2.8927 2.2425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8927 1.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3364 1.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7801 1.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7801 2.2425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3364 2.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2238 1.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6674 1.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6674 2.2425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2238 2.5637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2238 0.7781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0330 2.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6000 2.3600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1670 2.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1670 3.3420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6000 3.6693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0330 3.3420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3364 0.6368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7738 3.6923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4083 2.6962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0380 1.2367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1033 -0.7794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4321 -1.0885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2622 -0.4941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4321 0.1040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1033 0.4132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0665 -0.1813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7231 -0.1813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0342 -1.2926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3029 -1.5706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7908 -0.7993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7311 0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4303 -0.3934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0087 -0.2281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5907 -0.3934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8916 0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3131 -0.0377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1724 -0.0221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5513 0.3749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3257 -0.1230 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1423 -0.3934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4083 -1.3050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9942 -2.2946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2532 -2.7432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4385 -2.2946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1337 -2.8225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3246 -2.8225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0189 -2.2930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4074 -2.2930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1017 -2.8225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4074 -3.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0189 -3.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4303 -2.8225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4739 -2.8961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1027 -3.3861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5682 -3.1782 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0524 -3.1727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4272 -2.7978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8948 -3.0446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9877 -3.1928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6813 -3.4142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2619 -3.6923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2933 -2.2604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4964 -2.4739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
27 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
26 21 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
37 39 1 0 0 0 0
36 40 1 0 0 0 0
35 41 1 0 0 0 0
33 28 1 0 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
54 55 1 1 0 0 0
55 56 1 1 0 0 0
57 56 1 1 0 0 0
57 58 1 0 0 0 0
58 59 1 0 0 0 0
59 54 1 0 0 0 0
54 60 1 0 0 0 0
55 61 1 0 0 0 0
56 62 1 0 0 0 0
57 53 1 0 0 0 0
59 63 1 0 0 0 0
63 64 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 63 64
M SBL 1 1 69
M SMT 1 ^CH2OH
M SBV 1 69 -6.5251 7.4198
S SKP 8
ID FL5FAAGS0062
KNApSAcK_ID C00005885
NAME Kaempferol 3-(6-[4-glucosyl-p-coumaryl]glucosyl)(1->2)-rhamnoside
CAS_RN 142997-34-0
FORMULA C42H46O22
EXACTMASS 902.248073156
AVERAGEMASS 902.80144
SMILES CC(C7O)OC(C(C7O)OC(C(O)6)OC(C(O)C6O)COC(=O)C=Cc(c5)ccc(c5)OC(O4)C(C(O)C(O)C4CO)O)OC(C(=O)1)=C(c(c3)ccc(c3)O)Oc(c2)c1c(O)cc(O)2
M END