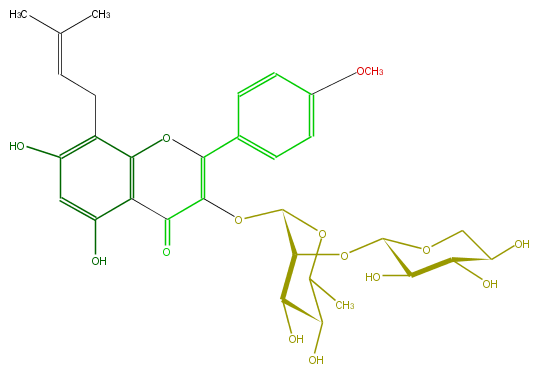
Mol:FL5FABGI0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -4.1944 0.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1944 0.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1944 -0.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1944 -0.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4799 -0.6611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4799 -0.6611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7654 -0.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7654 -0.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7654 0.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7654 0.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4799 0.9888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4799 0.9888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0510 -0.6611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0510 -0.6611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3365 -0.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3365 -0.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3365 0.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3365 0.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0510 0.9888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0510 0.9888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0510 -1.3044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0510 -1.3044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6223 0.9887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6223 0.9887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1059 0.5682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1059 0.5682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8340 0.9887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8340 0.9887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8340 1.8295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8340 1.8295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1059 2.2499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1059 2.2499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6223 1.8295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6223 1.8295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4799 -1.4859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4799 -1.4859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6223 -0.6610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6223 -0.6610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9789 0.8313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9789 0.8313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4799 1.8135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4799 1.8135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1941 2.2259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1941 2.2259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1941 3.0505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1941 3.0505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9083 3.4629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9083 3.4629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4799 3.4629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4799 3.4629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2469 -2.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2469 -2.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0537 -2.7364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0537 -2.7364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7978 -1.8406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7978 -1.8406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0537 -0.9394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0537 -0.9394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2469 -0.4734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2469 -0.4734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5029 -1.3693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5029 -1.3693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4922 -1.3693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4922 -1.3693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4542 -3.0440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4542 -3.0440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8591 -3.4629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8591 -3.4629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3894 -2.3638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3894 -2.3638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2523 -1.0382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2523 -1.0382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8205 -1.7882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8205 -1.7882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6387 -1.4701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6387 -1.4701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4282 -1.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4282 -1.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8545 -0.8877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8545 -0.8877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1387 -1.2655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1387 -1.2655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1425 -1.7915 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1425 -1.7915 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3357 -1.9284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3357 -1.9284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9789 -1.1416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9789 -1.1416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8301 2.3173 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8301 2.3173 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7477 1.7876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7477 1.7876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 6 21 1 0 0 0 0 | + | 6 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 26 1 1 0 0 0 | + | 31 26 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 26 33 1 0 0 0 0 | + | 26 33 1 0 0 0 0 |
| − | 27 34 1 0 0 0 0 | + | 27 34 1 0 0 0 0 |
| − | 28 35 1 0 0 0 0 | + | 28 35 1 0 0 0 0 |
| − | 30 19 1 0 0 0 0 | + | 30 19 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 38 1 1 0 0 0 | + | 37 38 1 1 0 0 0 |
| − | 39 38 1 1 0 0 0 | + | 39 38 1 1 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 37 42 1 0 0 0 0 | + | 37 42 1 0 0 0 0 |
| − | 38 43 1 0 0 0 0 | + | 38 43 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 39 44 1 0 0 0 0 | + | 39 44 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 15 45 1 0 0 0 0 | + | 15 45 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 45 46 | + | M SAL 1 2 45 46 |
| − | M SBL 1 1 50 | + | M SBL 1 1 50 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 50 -0.9961 -0.4878 | + | M SBV 1 50 -0.9961 -0.4878 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FABGI0003 | + | ID FL5FABGI0003 |
| − | FORMULA C32H38O14 | + | FORMULA C32H38O14 |
| − | EXACTMASS 646.226155924 | + | EXACTMASS 646.226155924 |
| − | AVERAGEMASS 646.63572 | + | AVERAGEMASS 646.63572 |
| − | SMILES C(O4)(C(OC(O5)C(O)C(C(O)C5)O)C(C(O)C(C)4)O)OC(C3=O)=C(Oc(c32)c(c(cc(O)2)O)CC=C(C)C)c(c1)ccc(OC)c1 | + | SMILES C(O4)(C(OC(O5)C(O)C(C(O)C5)O)C(C(O)C(C)4)O)OC(C3=O)=C(Oc(c32)c(c(cc(O)2)O)CC=C(C)C)c(c1)ccc(OC)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-4.1944 0.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1944 -0.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4799 -0.6611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7654 -0.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7654 0.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4799 0.9888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0510 -0.6611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3365 -0.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3365 0.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0510 0.9888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0510 -1.3044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6223 0.9887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1059 0.5682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8340 0.9887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8340 1.8295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1059 2.2499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6223 1.8295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4799 -1.4859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6223 -0.6610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9789 0.8313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4799 1.8135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1941 2.2259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1941 3.0505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9083 3.4629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4799 3.4629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2469 -2.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0537 -2.7364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7978 -1.8406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0537 -0.9394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2469 -0.4734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5029 -1.3693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4922 -1.3693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4542 -3.0440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8591 -3.4629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3894 -2.3638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2523 -1.0382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8205 -1.7882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6387 -1.4701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4282 -1.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8545 -0.8877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1387 -1.2655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1425 -1.7915 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3357 -1.9284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9789 -1.1416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8301 2.3173 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7477 1.7876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
8 19 1 0 0 0 0
1 20 1 0 0 0 0
6 21 1 0 0 0 0
21 22 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
23 25 1 0 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 1 0 0 0
31 26 1 1 0 0 0
31 32 1 0 0 0 0
26 33 1 0 0 0 0
27 34 1 0 0 0 0
28 35 1 0 0 0 0
30 19 1 0 0 0 0
36 37 1 1 0 0 0
37 38 1 1 0 0 0
39 38 1 1 0 0 0
39 40 1 0 0 0 0
40 41 1 0 0 0 0
41 36 1 0 0 0 0
37 42 1 0 0 0 0
38 43 1 0 0 0 0
32 36 1 0 0 0 0
39 44 1 0 0 0 0
45 46 1 0 0 0 0
15 45 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 45 46
M SBL 1 1 50
M SMT 1 OCH3
M SBV 1 50 -0.9961 -0.4878
S SKP 5
ID FL5FABGI0003
FORMULA C32H38O14
EXACTMASS 646.226155924
AVERAGEMASS 646.63572
SMILES C(O4)(C(OC(O5)C(O)C(C(O)C5)O)C(C(O)C(C)4)O)OC(C3=O)=C(Oc(c32)c(c(cc(O)2)O)CC=C(C)C)c(c1)ccc(OC)c1
M END