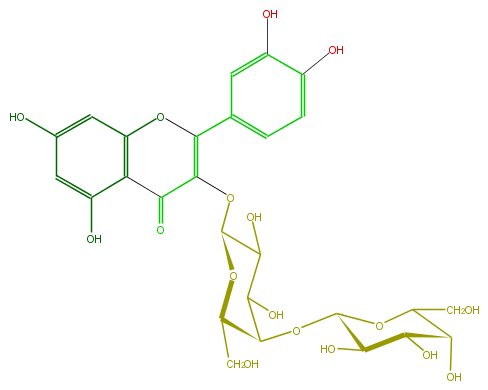
Mol:FL5FACGA0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -4.0473 1.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0473 1.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0473 0.3564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0473 0.3564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3462 -0.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3462 -0.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6451 0.3564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6451 0.3564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6451 1.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6451 1.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3462 1.5708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3462 1.5708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9440 -0.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9440 -0.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2429 0.3564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2429 0.3564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2429 1.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2429 1.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9440 1.5708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9440 1.5708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9440 -0.6796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9440 -0.6796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5420 1.5707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5420 1.5707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1725 1.1581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1725 1.1581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8871 1.5707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8871 1.5707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8871 2.3958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8871 2.3958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1725 2.8083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1725 2.8083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5420 2.3958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5420 2.3958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7482 1.5707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7482 1.5707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3462 -0.8577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3462 -0.8577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4974 2.9213 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4974 2.9213 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5420 -0.0482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5420 -0.0482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1725 3.6332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1725 3.6332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0373 -1.1933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0373 -1.1933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7428 -0.7428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7428 -0.7428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4953 -1.6091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4953 -1.6091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7428 -2.4805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7428 -2.4805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0373 -2.9311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0373 -2.9311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2101 -2.0648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2101 -2.0648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1428 -0.4214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1428 -0.4214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2853 -2.3816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2853 -2.3816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7679 -2.7010 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7679 -2.7010 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5623 -2.3777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5623 -2.3777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1198 -3.1137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1198 -3.1137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9226 -2.8015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9226 -2.8015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8596 -2.8579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8596 -2.8579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0740 -2.2934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0740 -2.2934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4320 -2.6008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4320 -2.6008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4802 -3.0763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4802 -3.0763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3783 -3.1789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3783 -3.1789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8425 -3.6332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8425 -3.6332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5668 -3.3627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5668 -3.3627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3021 -2.6275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3021 -2.6275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8478 -2.2967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8478 -2.2967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7482 -2.8166 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7482 -2.8166 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 24 21 1 0 0 0 0 | + | 24 21 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 35 40 1 0 0 0 0 | + | 35 40 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 26 41 1 0 0 0 0 | + | 26 41 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 36 43 1 0 0 0 0 | + | 36 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 46 -0.1760 0.8822 | + | M SBV 1 46 -0.1760 0.8822 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 48 -0.7738 0.0034 | + | M SBV 2 48 -0.7738 0.0034 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGA0005 | + | ID FL5FACGA0005 |
| − | FORMULA C27H30O17 | + | FORMULA C27H30O17 |
| − | EXACTMASS 626.148299534 | + | EXACTMASS 626.148299534 |
| − | AVERAGEMASS 626.5169000000001 | + | AVERAGEMASS 626.5169000000001 |
| − | SMILES c(c(O)5)cc(cc5O)C(=C1OC(C(O)3)OC(C(OC(O4)C(O)C(O)C(O)C4CO)C3O)CO)Oc(c2)c(c(O)cc2O)C1=O | + | SMILES c(c(O)5)cc(cc5O)C(=C1OC(C(O)3)OC(C(OC(O4)C(O)C(O)C(O)C4CO)C3O)CO)Oc(c2)c(c(O)cc2O)C1=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-4.0473 1.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0473 0.3564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3462 -0.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6451 0.3564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6451 1.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3462 1.5708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9440 -0.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2429 0.3564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2429 1.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9440 1.5708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9440 -0.6796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5420 1.5707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1725 1.1581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8871 1.5707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8871 2.3958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1725 2.8083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5420 2.3958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7482 1.5707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3462 -0.8577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4974 2.9213 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5420 -0.0482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1725 3.6332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0373 -1.1933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7428 -0.7428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4953 -1.6091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7428 -2.4805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0373 -2.9311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2101 -2.0648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1428 -0.4214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2853 -2.3816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7679 -2.7010 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5623 -2.3777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1198 -3.1137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9226 -2.8015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8596 -2.8579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0740 -2.2934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4320 -2.6008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4802 -3.0763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3783 -3.1789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8425 -3.6332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5668 -3.3627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3021 -2.6275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8478 -2.2967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7482 -2.8166 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
20 15 1 0 0 0 0
8 21 1 0 0 0 0
16 22 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
28 30 1 0 0 0 0
27 31 1 0 0 0 0
24 21 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
33 38 1 0 0 0 0
34 39 1 0 0 0 0
31 32 1 0 0 0 0
35 40 1 0 0 0 0
41 42 1 0 0 0 0
26 41 1 0 0 0 0
43 44 1 0 0 0 0
36 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 CH2OH
M SBV 1 46 -0.1760 0.8822
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 48
M SMT 2 CH2OH
M SBV 2 48 -0.7738 0.0034
S SKP 5
ID FL5FACGA0005
FORMULA C27H30O17
EXACTMASS 626.148299534
AVERAGEMASS 626.5169000000001
SMILES c(c(O)5)cc(cc5O)C(=C1OC(C(O)3)OC(C(OC(O4)C(O)C(O)C(O)C4CO)C3O)CO)Oc(c2)c(c(O)cc2O)C1=O
M END