Mol:FL5FACGS0027
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.2135 0.8620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2135 0.8620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2135 0.2197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2135 0.2197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6572 -0.1015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6572 -0.1015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1009 0.2197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1009 0.2197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1009 0.8620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1009 0.8620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6572 1.1832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6572 1.1832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5446 -0.1015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5446 -0.1015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9883 0.2197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9883 0.2197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9883 0.8620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9883 0.8620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5446 1.1832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5446 1.1832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5446 -0.6023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5446 -0.6023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2879 1.3069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2879 1.3069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2791 0.9795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2791 0.9795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8461 1.3069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8461 1.3069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8461 1.9615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8461 1.9615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2791 2.2889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2791 2.2889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2879 1.9615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2879 1.9615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6572 -0.7436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6572 -0.7436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4529 2.3119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4529 2.3119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7085 1.1832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7085 1.1832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4250 -0.1898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4250 -0.1898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2791 2.9434 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2791 2.9434 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2152 -1.4899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2152 -1.4899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8434 -1.8526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8434 -1.8526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6440 -1.1551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6440 -1.1551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8434 -0.4534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8434 -0.4534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2152 -0.0906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2152 -0.0906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4144 -0.7881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4144 -0.7881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1848 -0.7881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1848 -0.7881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3765 -2.0921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3765 -2.0921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6918 -2.4183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6918 -2.4183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2643 -1.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2643 -1.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7814 -2.0366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7814 -2.0366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4806 -2.5576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4806 -2.5576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0591 -2.3923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0591 -2.3923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6411 -2.5576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6411 -2.5576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1225 -2.0048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1225 -2.0048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3634 -2.2018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3634 -2.2018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2227 -2.1863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2227 -2.1863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6016 -1.7893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6016 -1.7893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9088 -1.2074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9088 -1.2074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9940 -2.5309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9940 -2.5309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7085 -2.9434 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7085 -2.9434 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 23 1 1 0 0 0 | + | 28 23 1 1 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 25 32 1 0 0 0 0 | + | 25 32 1 0 0 0 0 |
| − | 27 21 1 0 0 0 0 | + | 27 21 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 38 40 1 0 0 0 0 | + | 38 40 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 31 34 1 0 0 0 0 | + | 31 34 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 46 -8.3427 6.2100 | + | M SBV 1 46 -8.3427 6.2100 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGS0027 | + | ID FL5FACGS0027 |
| − | KNApSAcK_ID C00005416 | + | KNApSAcK_ID C00005416 |
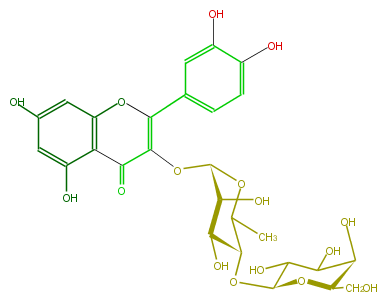
| − | NAME Quercetin 3-galactosyl-(1->4)-rhamnoside | + | NAME Quercetin 3-galactosyl-(1->4)-rhamnoside |
| − | CAS_RN 98618-77-0 | + | CAS_RN 98618-77-0 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES C(=C3OC(C(O)5)OC(C)C(C5O)OC(O4)C(O)C(O)C(C4CO)O)(Oc(c2)c(C3=O)c(O)cc2O)c(c1)ccc(O)c1O | + | SMILES C(=C3OC(C(O)5)OC(C)C(C5O)OC(O4)C(O)C(O)C(C4CO)O)(Oc(c2)c(C3=O)c(O)cc2O)c(c1)ccc(O)c1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-3.2135 0.8620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2135 0.2197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6572 -0.1015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1009 0.2197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1009 0.8620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6572 1.1832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5446 -0.1015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9883 0.2197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9883 0.8620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5446 1.1832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5446 -0.6023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2879 1.3069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2791 0.9795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8461 1.3069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8461 1.9615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2791 2.2889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2879 1.9615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6572 -0.7436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4529 2.3119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7085 1.1832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4250 -0.1898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2791 2.9434 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2152 -1.4899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8434 -1.8526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6440 -1.1551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8434 -0.4534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2152 -0.0906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4144 -0.7881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1848 -0.7881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3765 -2.0921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6918 -2.4183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2643 -1.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7814 -2.0366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4806 -2.5576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0591 -2.3923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6411 -2.5576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1225 -2.0048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3634 -2.2018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2227 -2.1863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6016 -1.7893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9088 -1.2074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9940 -2.5309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7085 -2.9434 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
21 8 1 0 0 0 0
4 3 1 0 0 0 0
16 22 1 0 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 1 0 0 0
28 23 1 1 0 0 0
28 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
25 32 1 0 0 0 0
27 21 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
33 39 1 0 0 0 0
38 40 1 0 0 0 0
37 41 1 0 0 0 0
31 34 1 0 0 0 0
36 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 46
M SMT 1 CH2OH
M SBV 1 46 -8.3427 6.2100
S SKP 8
ID FL5FACGS0027
KNApSAcK_ID C00005416
NAME Quercetin 3-galactosyl-(1->4)-rhamnoside
CAS_RN 98618-77-0
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES C(=C3OC(C(O)5)OC(C)C(C5O)OC(O4)C(O)C(O)C(C4CO)O)(Oc(c2)c(C3=O)c(O)cc2O)c(c1)ccc(O)c1O
M END