Mol:FL5FACGS0095
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | 5.2475 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2475 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5330 0.2558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5330 0.2558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5330 1.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5330 1.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2475 1.4933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2475 1.4933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9619 1.0809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9619 1.0809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9619 0.2559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9619 0.2559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8185 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8185 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1040 0.2558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1040 0.2558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3896 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3896 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3896 -0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3896 -0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1040 -1.3941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1040 -1.3941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8185 -0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8185 -0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6751 0.2558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6751 0.2558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9606 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9606 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9606 -0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9606 -0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6751 -1.3942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6751 -1.3942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1040 -2.0549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1040 -2.0549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2462 0.2558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2462 0.2558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4175 -1.4272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4175 -1.4272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6751 -2.2192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6751 -2.2192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5538 1.4226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5538 1.4226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2475 2.2192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2475 2.2192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6440 -0.0476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6440 -0.0476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0538 -0.6245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0538 -0.6245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3415 -0.2077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3415 -0.2077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5162 -0.2121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5162 -0.2121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1063 0.3648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1063 0.3648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8187 -0.0518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8187 -0.0518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1279 0.4699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1279 0.4699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9789 -0.6432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9789 -0.6432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5558 -0.4961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5558 -0.4961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3340 -0.8702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3340 -0.8702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3745 0.2639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3745 0.2639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1812 1.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1812 1.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7792 1.6343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7792 1.6343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5704 1.4006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5704 1.4006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7637 0.5985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7637 0.5985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1657 0.0302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1657 0.0302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3586 -0.7708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3586 -0.7708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.5538 0.3651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.5538 0.3651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1676 1.9682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1676 1.9682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3911 1.2993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3911 1.2993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1981 2.1003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1981 2.1003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7939 0.7317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7939 0.7317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 5 21 1 0 0 0 0 | + | 5 21 1 0 0 0 0 |
| − | 4 22 1 0 0 0 0 | + | 4 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 25 32 1 0 0 0 0 | + | 25 32 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 36 41 1 0 0 0 0 | + | 36 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 44 29 1 0 0 0 0 | + | 44 29 1 0 0 0 0 |
| − | 26 18 1 0 0 0 0 | + | 26 18 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGS0095 | + | ID FL5FACGS0095 |
| − | KNApSAcK_ID C00013867 | + | KNApSAcK_ID C00013867 |
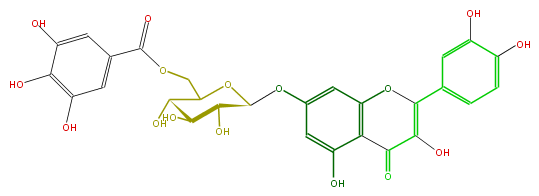
| − | NAME Quercetin 7-(6''-galloylglucoside);2-(3,4-Dihydroxyphenyl)-3,5-dihydroxy-7-[[6-O-(3,4,5-trihydroxybenzoyl)-beta-D-glucopyranosyl]oxy]-4H-1-benzopyran-4-one | + | NAME Quercetin 7-(6''-galloylglucoside);2-(3,4-Dihydroxyphenyl)-3,5-dihydroxy-7-[[6-O-(3,4,5-trihydroxybenzoyl)-beta-D-glucopyranosyl]oxy]-4H-1-benzopyran-4-one |
| − | CAS_RN 231289-25-1 | + | CAS_RN 231289-25-1 |
| − | FORMULA C28H24O16 | + | FORMULA C28H24O16 |
| − | EXACTMASS 616.1064347199999 | + | EXACTMASS 616.1064347199999 |
| − | AVERAGEMASS 616.48056 | + | AVERAGEMASS 616.48056 |
| − | SMILES c(c1OC(O4)C(C(C(O)C(COC(=O)c(c5)cc(O)c(c(O)5)O)4)O)O)c(O2)c(C(=O)C(O)=C(c(c3)ccc(c3O)O)2)c(c1)O | + | SMILES c(c1OC(O4)C(C(C(O)C(COC(=O)c(c5)cc(O)c(c(O)5)O)4)O)O)c(O2)c(C(=O)C(O)=C(c(c3)ccc(c3O)O)2)c(c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
5.2475 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5330 0.2558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5330 1.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2475 1.4933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9619 1.0809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9619 0.2559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8185 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1040 0.2558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3896 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3896 -0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1040 -1.3941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8185 -0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6751 0.2558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9606 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9606 -0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6751 -1.3942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1040 -2.0549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2462 0.2558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4175 -1.4272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6751 -2.2192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.5538 1.4226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2475 2.2192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6440 -0.0476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0538 -0.6245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3415 -0.2077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5162 -0.2121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1063 0.3648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8187 -0.0518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1279 0.4699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9789 -0.6432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5558 -0.4961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3340 -0.8702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3745 0.2639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1812 1.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7792 1.6343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5704 1.4006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7637 0.5985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1657 0.0302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3586 -0.7708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.5538 0.3651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.1676 1.9682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3911 1.2993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1981 2.1003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7939 0.7317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
16 20 1 0 0 0 0
5 21 1 0 0 0 0
4 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
28 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
25 32 1 0 0 0 0
33 34 2 0 0 0 0
34 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 33 1 0 0 0 0
38 39 1 0 0 0 0
37 40 1 0 0 0 0
36 41 1 0 0 0 0
34 42 1 0 0 0 0
42 43 2 0 0 0 0
42 44 1 0 0 0 0
44 29 1 0 0 0 0
26 18 1 0 0 0 0
S SKP 8
ID FL5FACGS0095
KNApSAcK_ID C00013867
NAME Quercetin 7-(6''-galloylglucoside);2-(3,4-Dihydroxyphenyl)-3,5-dihydroxy-7-[[6-O-(3,4,5-trihydroxybenzoyl)-beta-D-glucopyranosyl]oxy]-4H-1-benzopyran-4-one
CAS_RN 231289-25-1
FORMULA C28H24O16
EXACTMASS 616.1064347199999
AVERAGEMASS 616.48056
SMILES c(c1OC(O4)C(C(C(O)C(COC(=O)c(c5)cc(O)c(c(O)5)O)4)O)O)c(O2)c(C(=O)C(O)=C(c(c3)ccc(c3O)O)2)c(c1)O
M END