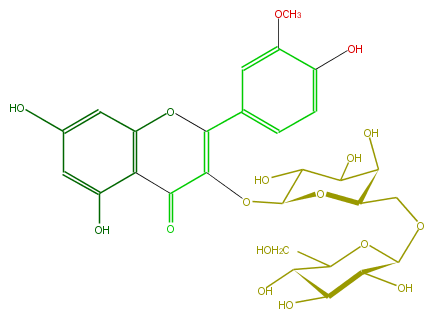
Mol:FL5FADGA0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.1489 -0.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1489 -0.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1489 -0.8689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1489 -0.8689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4344 -1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4344 -1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7199 -0.8689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7199 -0.8689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7199 -0.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7199 -0.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4344 0.3685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4344 0.3685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0055 -1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0055 -1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2910 -0.8689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2910 -0.8689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2910 -0.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2910 -0.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0055 0.3685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0055 0.3685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0055 -1.9750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0055 -1.9750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4230 0.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4230 0.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1511 -0.0581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1511 -0.0581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8793 0.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8793 0.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8793 1.2031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8793 1.2031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1511 1.6236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1511 1.6236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4230 1.2031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4230 1.2031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4344 -1.9963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4344 -1.9963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6139 1.6272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6139 1.6272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0023 0.4488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0023 0.4488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6311 -0.7947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6311 -0.7947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2357 -1.4792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2357 -1.4792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9958 -1.2620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9958 -1.2620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7605 -1.4792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7605 -1.4792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1558 -0.7947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1558 -0.7947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3956 -1.0119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3956 -1.0119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8969 -0.9914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8969 -0.9914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5959 -0.5547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5959 -0.5547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9303 -0.0530 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9303 -0.0530 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5085 -1.3582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5085 -1.3582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4428 -2.8019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4428 -2.8019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1473 -3.3493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1473 -3.3493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8185 -2.8572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8185 -2.8572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5394 -2.6556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5394 -2.6556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8732 -2.2709 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8732 -2.2709 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1815 -2.7509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1815 -2.7509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8214 -3.2129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8214 -3.2129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3873 -3.4886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3873 -3.4886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6101 -3.1576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6101 -3.1576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0023 -1.9138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0023 -1.9138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5081 -1.4386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5081 -1.4386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1511 2.3362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1511 2.3362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7180 3.4886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7180 3.4886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4203 -2.3913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4203 -2.3913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5804 -2.9419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5804 -2.9419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 40 30 1 0 0 0 0 | + | 40 30 1 0 0 0 0 |
| − | 22 41 1 0 0 0 0 | + | 22 41 1 0 0 0 0 |
| − | 41 8 1 0 0 0 0 | + | 41 8 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 16 42 1 0 0 0 0 | + | 16 42 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 36 44 1 0 0 0 0 | + | 36 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 47 0.0000 -0.7126 | + | M SBV 1 47 0.0000 -0.7126 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 44 45 | + | M SAL 2 2 44 45 |
| − | M SBL 2 1 49 | + | M SBL 2 1 49 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 49 0.7612 -0.3596 | + | M SBV 2 49 0.7612 -0.3596 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGA0005 | + | ID FL5FADGA0005 |
| − | FORMULA C28H32O17 | + | FORMULA C28H32O17 |
| − | EXACTMASS 640.163949598 | + | EXACTMASS 640.163949598 |
| − | AVERAGEMASS 640.54348 | + | AVERAGEMASS 640.54348 |
| − | SMILES O=C(c23)C(OC(C(O)4)OC(COC(C(O)5)OC(CO)C(O)C5O)C(O)C4O)=C(Oc2cc(O)cc3O)c(c1)cc(OC)c(O)c1 | + | SMILES O=C(c23)C(OC(C(O)4)OC(COC(C(O)5)OC(CO)C(O)C5O)C(O)C4O)=C(Oc2cc(O)cc3O)c(c1)cc(OC)c(O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-3.1489 -0.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1489 -0.8689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4344 -1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7199 -0.8689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7199 -0.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4344 0.3685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0055 -1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2910 -0.8689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2910 -0.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0055 0.3685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0055 -1.9750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4230 0.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1511 -0.0581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8793 0.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8793 1.2031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1511 1.6236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4230 1.2031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4344 -1.9963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6139 1.6272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0023 0.4488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6311 -0.7947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2357 -1.4792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9958 -1.2620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7605 -1.4792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1558 -0.7947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3956 -1.0119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8969 -0.9914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5959 -0.5547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9303 -0.0530 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5085 -1.3582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4428 -2.8019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1473 -3.3493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8185 -2.8572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5394 -2.6556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8732 -2.2709 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1815 -2.7509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8214 -3.2129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3873 -3.4886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6101 -3.1576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0023 -1.9138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5081 -1.4386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1511 2.3362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7180 3.4886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4203 -2.3913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5804 -2.9419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
40 30 1 0 0 0 0
22 41 1 0 0 0 0
41 8 1 0 0 0 0
42 43 1 0 0 0 0
16 42 1 0 0 0 0
44 45 1 0 0 0 0
36 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 47
M SMT 1 OCH3
M SBV 1 47 0.0000 -0.7126
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 44 45
M SBL 2 1 49
M SMT 2 ^ CH2OH
M SBV 2 49 0.7612 -0.3596
S SKP 5
ID FL5FADGA0005
FORMULA C28H32O17
EXACTMASS 640.163949598
AVERAGEMASS 640.54348
SMILES O=C(c23)C(OC(C(O)4)OC(COC(C(O)5)OC(CO)C(O)C5O)C(O)C4O)=C(Oc2cc(O)cc3O)c(c1)cc(OC)c(O)c1
M END