Mol:FL5FAGGS0030
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | 3.1468 1.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1468 1.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4323 1.4932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4323 1.4932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4323 2.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4323 2.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1468 2.7307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1468 2.7307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8612 2.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8612 2.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8612 1.4932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8612 1.4932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7178 1.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7178 1.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0034 1.4932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0034 1.4932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2889 1.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2889 1.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2889 0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2889 0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0034 -0.1568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0034 -0.1568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7178 0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7178 0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4256 1.4932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4256 1.4932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1401 1.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1401 1.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1401 0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1401 0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4256 -0.1568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4256 -0.1568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0034 -0.8638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0034 -0.8638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8545 1.4932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8545 1.4932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4323 -0.1568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4323 -0.1568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4256 -0.8478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4256 -0.8478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5757 2.7307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5757 2.7307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1468 3.5557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1468 3.5557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5392 1.1018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5392 1.1018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4218 -2.5150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4218 -2.5150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9149 -2.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9149 -2.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6788 -2.3999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6788 -2.3999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1818 -1.7456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1818 -1.7456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9479 -1.4389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9479 -1.4389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1840 -1.1266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1840 -1.1266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6810 -1.7809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6810 -1.7809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1981 -1.4140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1981 -1.4140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8289 -2.7656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8289 -2.7656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1655 -2.4700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1655 -2.4700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5053 -1.5803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5053 -1.5803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0421 -2.1277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0421 -2.1277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3640 -2.8311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3640 -2.8311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8660 -2.1277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8660 -2.1277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2779 -2.8412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2779 -2.8412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1018 -2.8412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1018 -2.8412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5143 -2.1267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5143 -2.1267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3393 -2.1267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3393 -2.1267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7518 -2.8412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7518 -2.8412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3393 -3.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3393 -3.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5143 -3.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5143 -3.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5757 -2.8412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5757 -2.8412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 5 21 1 0 0 0 0 | + | 5 21 1 0 0 0 0 |
| − | 4 22 1 0 0 0 0 | + | 4 22 1 0 0 0 0 |
| − | 6 23 1 0 0 0 0 | + | 6 23 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 28 27 1 1 0 0 0 | + | 28 27 1 1 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 25 1 0 0 0 0 | + | 30 25 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 25 32 1 0 0 0 0 | + | 25 32 1 0 0 0 0 |
| − | 26 33 1 0 0 0 0 | + | 26 33 1 0 0 0 0 |
| − | 28 19 1 0 0 0 0 | + | 28 19 1 0 0 0 0 |
| − | 24 27 1 0 0 0 0 | + | 24 27 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 44 39 1 0 0 0 0 | + | 44 39 1 0 0 0 0 |
| − | 42 45 1 0 0 0 0 | + | 42 45 1 0 0 0 0 |
| − | 34 31 1 0 0 0 0 | + | 34 31 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAGGS0030 | + | ID FL5FAGGS0030 |
| − | KNApSAcK_ID C00013972 | + | KNApSAcK_ID C00013972 |
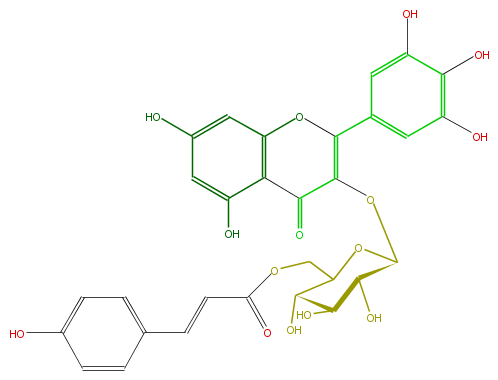
| − | NAME Myricetin 3-(6''-p-coumaroylglucoside) | + | NAME Myricetin 3-(6''-p-coumaroylglucoside) |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C30H26O15 | + | FORMULA C30H26O15 |
| − | EXACTMASS 626.127170162 | + | EXACTMASS 626.127170162 |
| − | AVERAGEMASS 626.51844 | + | AVERAGEMASS 626.51844 |
| − | SMILES C(c(c5)ccc(O)c5)=CC(OCC(C4O)OC(C(C(O)4)O)OC(=C2c(c3)cc(c(O)c(O)3)O)C(=O)c(c(O2)1)c(O)cc(O)c1)=O | + | SMILES C(c(c5)ccc(O)c5)=CC(OCC(C4O)OC(C(C(O)4)O)OC(=C2c(c3)cc(c(O)c(O)3)O)C(=O)c(c(O2)1)c(O)cc(O)c1)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
3.1468 1.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4323 1.4932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4323 2.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1468 2.7307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8612 2.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8612 1.4932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7178 1.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0034 1.4932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2889 1.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2889 0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0034 -0.1568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7178 0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4256 1.4932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1401 1.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1401 0.2557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4256 -0.1568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0034 -0.8638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8545 1.4932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4323 -0.1568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4256 -0.8478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5757 2.7307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1468 3.5557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5392 1.1018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4218 -2.5150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9149 -2.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6788 -2.3999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1818 -1.7456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9479 -1.4389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1840 -1.1266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6810 -1.7809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1981 -1.4140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8289 -2.7656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1655 -2.4700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5053 -1.5803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0421 -2.1277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3640 -2.8311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8660 -2.1277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2779 -2.8412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1018 -2.8412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5143 -2.1267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3393 -2.1267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7518 -2.8412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3393 -3.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5143 -3.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5757 -2.8412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
16 20 1 0 0 0 0
5 21 1 0 0 0 0
4 22 1 0 0 0 0
6 23 1 0 0 0 0
25 26 1 1 0 0 0
26 27 1 1 0 0 0
28 27 1 1 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
30 25 1 0 0 0 0
30 31 1 0 0 0 0
25 32 1 0 0 0 0
26 33 1 0 0 0 0
28 19 1 0 0 0 0
24 27 1 0 0 0 0
34 35 1 0 0 0 0
35 36 2 0 0 0 0
35 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
42 43 1 0 0 0 0
43 44 2 0 0 0 0
44 39 1 0 0 0 0
42 45 1 0 0 0 0
34 31 1 0 0 0 0
S SKP 8
ID FL5FAGGS0030
KNApSAcK_ID C00013972
NAME Myricetin 3-(6''-p-coumaroylglucoside)
CAS_RN -
FORMULA C30H26O15
EXACTMASS 626.127170162
AVERAGEMASS 626.51844
SMILES C(c(c5)ccc(O)c5)=CC(OCC(C4O)OC(C(C(O)4)O)OC(=C2c(c3)cc(c(O)c(O)3)O)C(=O)c(c(O2)1)c(O)cc(O)c1)=O
M END