Mol:FL5FAIGA0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 50 54 0 0 0 0 0 0 0 0999 V2000 | + | 50 54 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.9862 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9862 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9862 -0.6567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9862 -0.6567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4299 -0.9779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4299 -0.9779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8736 -0.6567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8736 -0.6567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8736 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8736 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4299 0.3068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4299 0.3068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3173 -0.9779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3173 -0.9779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7610 -0.6567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7610 -0.6567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7610 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7610 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3173 0.3068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3173 0.3068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3173 -1.4788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3173 -1.4788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0606 0.4304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0606 0.4304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4936 0.1031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4936 0.1031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0734 0.4304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0734 0.4304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0734 1.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0734 1.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4936 1.4125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4936 1.4125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0606 1.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0606 1.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6507 0.3693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6507 0.3693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4299 -1.5407 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4299 -1.5407 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9394 1.5851 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9394 1.5851 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6377 -0.5652 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -0.6377 -0.5652 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -0.2360 -1.0955 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.2360 -1.0955 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.3425 -0.8705 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.3425 -0.8705 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.9006 -0.8645 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.9006 -0.8645 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.4950 -0.4588 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.4950 -0.4588 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.0110 -0.7259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0110 -0.7259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1938 -0.8863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1938 -0.8863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3687 -1.7081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3687 -1.7081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6739 -1.4269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6739 -1.4269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4629 -0.5300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4629 -0.5300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1094 -1.6435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1094 -1.6435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1871 -0.3955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1871 -0.3955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0366 -0.1650 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.0366 -0.1650 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.6569 -0.6176 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.6569 -0.6176 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.2445 -0.5564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2445 -0.5564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7790 -0.8155 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.7790 -0.8155 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.1589 -0.3629 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.1589 -0.3629 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.5713 -0.4241 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.5713 -0.4241 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.9381 0.3942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9381 0.3942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6923 0.0277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6923 0.0277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5621 -0.0806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5621 -0.0806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2880 -1.0008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2880 -1.0008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2410 -1.5380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2410 -1.5380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7962 -1.9113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7962 -1.9113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4816 -2.0928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4816 -2.0928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0544 -2.4651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0544 -2.4651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2102 1.9886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2102 1.9886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2312 2.8859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2312 2.8859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0734 0.4304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0734 0.4304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0734 0.4304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0734 0.4304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 19 3 1 0 0 0 0 | + | 19 3 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 25 30 1 0 0 0 0 | + | 25 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 27 8 1 0 0 0 0 | + | 27 8 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 38 40 1 0 0 0 0 | + | 38 40 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 34 32 1 0 0 0 0 | + | 34 32 1 0 0 0 0 |
| − | 32 30 1 0 0 0 0 | + | 32 30 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 16 47 1 0 0 0 0 | + | 16 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 14 49 1 0 0 0 0 | + | 14 49 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 49 50 | + | M SAL 2 2 49 50 |
| − | M SBL 2 1 53 | + | M SBL 2 1 53 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 53 0.283 0.2242 | + | M SVB 2 53 0.283 0.2242 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 47 48 | + | M SAL 1 2 47 48 |
| − | M SBL 1 1 51 | + | M SBL 1 1 51 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 51 -0.2102 1.9886 | + | M SVB 1 51 -0.2102 1.9886 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAIGA0004 | + | ID FL5FAIGA0004 |
| − | KNApSAcK_ID C00006052 | + | KNApSAcK_ID C00006052 |
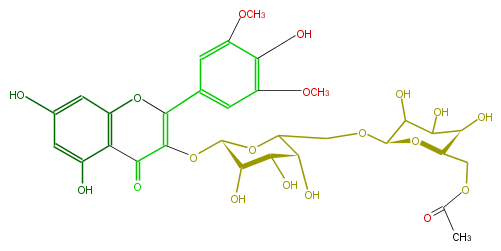
| − | NAME Syringetin 3-(,6'''-acetylglucosyl)(1->6)-galactoside | + | NAME Syringetin 3-(,6'''-acetylglucosyl)(1->6)-galactoside |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C31H36O19 | + | FORMULA C31H36O19 |
| − | EXACTMASS 712.18507897 | + | EXACTMASS 712.18507897 |
| − | AVERAGEMASS 712.6061400000001 | + | AVERAGEMASS 712.6061400000001 |
| − | SMILES C(c34)(C(=C(c(c5)cc(OC)c(O)c5OC)Oc3cc(O)cc(O)4)O[C@@H](O1)[C@@H]([C@H](O)[C@H](O)C1CO[C@H](O2)C(C(O)[C@@H](O)[C@H]2COC(C)=O)O)O)=O | + | SMILES C(c34)(C(=C(c(c5)cc(OC)c(O)c5OC)Oc3cc(O)cc(O)4)O[C@@H](O1)[C@@H]([C@H](O)[C@H](O)C1CO[C@H](O2)C(C(O)[C@@H](O)[C@H]2COC(C)=O)O)O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
50 54 0 0 0 0 0 0 0 0999 V2000
-3.9862 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9862 -0.6567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4299 -0.9779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8736 -0.6567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8736 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4299 0.3068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3173 -0.9779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7610 -0.6567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7610 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3173 0.3068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3173 -1.4788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0606 0.4304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4936 0.1031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0734 0.4304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0734 1.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4936 1.4125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0606 1.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6507 0.3693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4299 -1.5407 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9394 1.5851 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6377 -0.5652 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.2360 -1.0955 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.3425 -0.8705 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.9006 -0.8645 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.4950 -0.4588 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.0110 -0.7259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1938 -0.8863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3687 -1.7081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6739 -1.4269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4629 -0.5300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1094 -1.6435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1871 -0.3955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0366 -0.1650 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.6569 -0.6176 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.2445 -0.5564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7790 -0.8155 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.1589 -0.3629 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.5713 -0.4241 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.9381 0.3942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6923 0.0277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5621 -0.0806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2880 -1.0008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2410 -1.5380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7962 -1.9113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4816 -2.0928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0544 -2.4651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2102 1.9886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2312 2.8859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0734 0.4304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0734 0.4304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
4 3 1 0 0 0 0
1 18 1 0 0 0 0
19 3 1 0 0 0 0
15 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
25 30 1 0 0 0 0
24 31 1 0 0 0 0
27 8 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
33 39 1 0 0 0 0
38 40 1 0 0 0 0
37 41 1 0 0 0 0
36 42 1 0 0 0 0
34 32 1 0 0 0 0
32 30 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 2 0 0 0 0
44 46 1 0 0 0 0
16 47 1 0 0 0 0
47 48 1 0 0 0 0
14 49 1 0 0 0 0
49 50 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 49 50
M SBL 2 1 53
M SMT 2 OCH3
M SVB 2 53 0.283 0.2242
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 47 48
M SBL 1 1 51
M SMT 1 OCH3
M SVB 1 51 -0.2102 1.9886
S SKP 8
ID FL5FAIGA0004
KNApSAcK_ID C00006052
NAME Syringetin 3-(,6'''-acetylglucosyl)(1->6)-galactoside
CAS_RN -
FORMULA C31H36O19
EXACTMASS 712.18507897
AVERAGEMASS 712.6061400000001
SMILES C(c34)(C(=C(c(c5)cc(OC)c(O)c5OC)Oc3cc(O)cc(O)4)O[C@@H](O1)[C@@H]([C@H](O)[C@H](O)C1CO[C@H](O2)C(C(O)[C@@H](O)[C@H]2COC(C)=O)O)O)=O
M END