Mol:FL5FAJGS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.8044 0.6155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8044 0.6155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8044 -0.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8044 -0.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0899 -0.6222 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0899 -0.6222 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3754 -0.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3754 -0.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3754 0.6155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3754 0.6155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0899 1.0279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0899 1.0279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3391 -0.6222 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3391 -0.6222 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0536 -0.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0536 -0.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0536 0.6155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0536 0.6155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3391 1.0279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3391 1.0279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3391 -1.2654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3391 -1.2654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7679 1.0278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7679 1.0278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4961 0.6074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4961 0.6074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2244 1.0278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2244 1.0278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2244 1.8687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2244 1.8687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4961 2.2892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4961 2.2892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7679 1.8687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7679 1.8687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5187 1.0278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5187 1.0278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8294 -0.6280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8294 -0.6280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4961 3.1298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4961 3.1298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0899 -1.4469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0899 -1.4469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8602 1.8485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8602 1.8485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6285 1.8485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6285 1.8485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0762 1.3144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0762 1.3144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8874 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8874 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1191 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1191 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6712 1.0993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6712 1.0993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8701 1.5399 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8701 1.5399 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6933 2.3834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6933 2.3834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2303 2.2187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2303 2.2187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4954 0.3040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4954 0.3040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9524 0.6075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9524 0.6075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5845 -2.0619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5845 -2.0619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2721 -2.4589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2721 -2.4589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0539 -1.6954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0539 -1.6954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2721 -0.9274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2721 -0.9274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5845 -0.5302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5845 -0.5302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8027 -1.2938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8027 -1.2938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6459 -1.2938 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6459 -1.2938 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6388 -2.7638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6388 -2.7638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2061 -3.1298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2061 -3.1298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5280 -2.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5280 -2.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9524 2.2891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9524 2.2891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8701 1.7592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8701 1.7592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 18 26 1 0 0 0 0 | + | 18 26 1 0 0 0 0 |
| − | 14 32 1 0 0 0 0 | + | 14 32 1 0 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 1 0 0 0 | + | 37 38 1 1 0 0 0 |
| − | 38 33 1 1 0 0 0 | + | 38 33 1 1 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 34 41 1 0 0 0 0 | + | 34 41 1 0 0 0 0 |
| − | 35 42 1 0 0 0 0 | + | 35 42 1 0 0 0 0 |
| − | 37 19 1 0 0 0 0 | + | 37 19 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 15 43 1 0 0 0 0 | + | 15 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 48 | + | M SBL 1 1 48 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 48 -0.7280 -0.4204 | + | M SBV 1 48 -0.7280 -0.4204 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAJGS0002 | + | ID FL5FAJGS0002 |
| − | FORMULA C28H32O16 | + | FORMULA C28H32O16 |
| − | EXACTMASS 624.1690349759999 | + | EXACTMASS 624.1690349759999 |
| − | AVERAGEMASS 624.54408 | + | AVERAGEMASS 624.54408 |
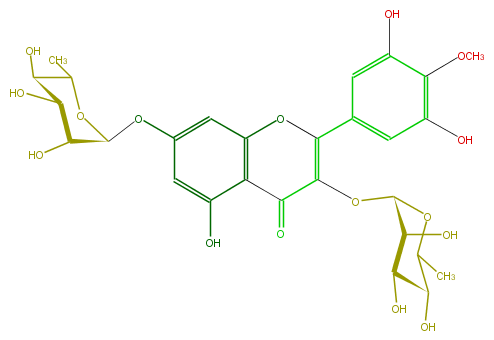
| − | SMILES OC(C1Oc(c5)cc(c(c53)C(=O)C(=C(c(c4)cc(c(OC)c4O)O)O3)OC(O2)C(O)C(O)C(C(C)2)O)O)C(C(C(O1)C)O)O | + | SMILES OC(C1Oc(c5)cc(c(c53)C(=O)C(=C(c(c4)cc(c(OC)c4O)O)O3)OC(O2)C(O)C(O)C(C(C)2)O)O)C(C(C(O1)C)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-1.8044 0.6155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8044 -0.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0899 -0.6222 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3754 -0.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3754 0.6155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0899 1.0279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3391 -0.6222 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0536 -0.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0536 0.6155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3391 1.0279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3391 -1.2654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7679 1.0278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4961 0.6074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2244 1.0278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2244 1.8687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4961 2.2892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7679 1.8687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5187 1.0278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8294 -0.6280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4961 3.1298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0899 -1.4469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8602 1.8485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6285 1.8485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0762 1.3144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8874 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1191 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6712 1.0993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8701 1.5399 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6933 2.3834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2303 2.2187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4954 0.3040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9524 0.6075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5845 -2.0619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2721 -2.4589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0539 -1.6954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2721 -0.9274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5845 -0.5302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8027 -1.2938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6459 -1.2938 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6388 -2.7638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2061 -3.1298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5280 -2.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9524 2.2891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8701 1.7592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
16 20 1 0 0 0 0
3 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
24 28 1 0 0 0 0
23 29 1 0 0 0 0
22 30 1 0 0 0 0
25 31 1 0 0 0 0
18 26 1 0 0 0 0
14 32 1 0 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 38 1 1 0 0 0
38 33 1 1 0 0 0
38 39 1 0 0 0 0
33 40 1 0 0 0 0
34 41 1 0 0 0 0
35 42 1 0 0 0 0
37 19 1 0 0 0 0
43 44 1 0 0 0 0
15 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 48
M SMT 1 OCH3
M SBV 1 48 -0.7280 -0.4204
S SKP 5
ID FL5FAJGS0002
FORMULA C28H32O16
EXACTMASS 624.1690349759999
AVERAGEMASS 624.54408
SMILES OC(C1Oc(c5)cc(c(c53)C(=O)C(=C(c(c4)cc(c(OC)c4O)O)O3)OC(O2)C(O)C(O)C(C(C)2)O)O)C(C(C(O1)C)O)O
M END