Mol:FL5FCDGA0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.3586 -4.2877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3586 -4.2877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7381 -4.4539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7381 -4.4539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2839 -3.9997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2839 -3.9997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4501 -3.3792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4501 -3.3792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0707 -3.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0707 -3.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5249 -3.6672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5249 -3.6672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9959 -2.9250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9959 -2.9250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1622 -2.3046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1622 -2.3046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7827 -2.1383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7827 -2.1383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2369 -2.5925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2369 -2.5925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5122 -3.0545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5122 -3.0545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0310 -1.3465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0310 -1.3465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5681 -0.8836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5681 -0.8836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7376 -0.2513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7376 -0.2513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3700 -0.0818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3700 -0.0818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8328 -0.5447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8328 -0.5447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6634 -1.1771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6634 -1.1771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6637 -4.1658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6637 -4.1658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1111 0.8841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1111 0.8841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6184 -1.9906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6184 -1.9906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0837 -0.9410 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.0837 -0.9410 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.3791 -1.4526 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.3791 -1.4526 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.1890 -1.2902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1890 -1.2902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7604 -1.4526 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.7604 -1.4526 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.0559 -0.9410 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.0559 -0.9410 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.4878 -1.1033 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.4878 -1.1033 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.2778 -0.4074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2778 -0.4074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5285 -0.6372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5285 -0.6372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6237 1.7063 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.6237 1.7063 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -2.2848 1.2223 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.2848 1.2223 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1919 1.8057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1919 1.8057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8032 2.2550 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.8032 2.2550 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1421 2.7391 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1421 2.7391 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.2351 2.1556 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.2351 2.1556 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.1894 1.7557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1894 1.7557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6402 2.3895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6402 2.3895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8479 3.0895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8479 3.0895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3686 -0.6786 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3686 -0.6786 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0487 -4.4726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0487 -4.4726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0146 -4.7314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0146 -4.7314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2308 -1.4371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2308 -1.4371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9452 -1.8496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9452 -1.8496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3160 -0.1218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3160 -0.1218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0686 0.5367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0686 0.5367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6185 2.6879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6185 2.6879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9427 3.1610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9427 3.1610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 20 22 1 0 0 0 0 | + | 20 22 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 30 19 1 0 0 0 0 | + | 30 19 1 0 0 0 0 |
| − | 25 38 1 0 0 0 0 | + | 25 38 1 0 0 0 0 |
| − | 1 39 1 0 0 0 0 | + | 1 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 24 41 1 0 0 0 0 | + | 24 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 16 43 1 0 0 0 0 | + | 16 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 32 45 1 0 0 0 0 | + | 32 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 45 46 | + | M SAL 4 2 45 46 |
| − | M SBL 4 1 49 | + | M SBL 4 1 49 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 49 3.2727 1.141 | + | M SVB 4 49 3.2727 1.141 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 41 42 | + | M SAL 3 2 41 42 |
| − | M SBL 3 1 45 | + | M SBL 3 1 45 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 45 1.2308 -1.4371 | + | M SVB 3 45 1.2308 -1.4371 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 47 0.1462 1.7287 | + | M SVB 2 47 0.1462 1.7287 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 43 -3.9871 0.3444 | + | M SVB 1 43 -3.9871 0.3444 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FCDGA0002 | + | ID FL5FCDGA0002 |
| − | KNApSAcK_ID C00005610 | + | KNApSAcK_ID C00005610 |
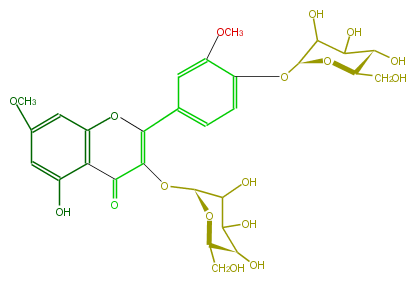
| − | NAME Rhamnazin 3-galactoside-4'-glucoside | + | NAME Rhamnazin 3-galactoside-4'-glucoside |
| − | CAS_RN 106009-50-1 | + | CAS_RN 106009-50-1 |
| − | FORMULA C29H34O17 | + | FORMULA C29H34O17 |
| − | EXACTMASS 654.179599662 | + | EXACTMASS 654.179599662 |
| − | AVERAGEMASS 654.57006 | + | AVERAGEMASS 654.57006 |
| − | SMILES C(C5O)([C@@H](O[C@@H]([C@@H]5O)CO)Oc(c4OC)ccc(c4)C(=C2O[C@H](O3)C(O)C([C@@H](O)[C@H]3CO)O)Oc(c1C2=O)cc(cc1O)OC)O | + | SMILES C(C5O)([C@@H](O[C@@H]([C@@H]5O)CO)Oc(c4OC)ccc(c4)C(=C2O[C@H](O3)C(O)C([C@@H](O)[C@H]3CO)O)Oc(c1C2=O)cc(cc1O)OC)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-2.3586 -4.2877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7381 -4.4539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2839 -3.9997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4501 -3.3792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0707 -3.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5249 -3.6672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9959 -2.9250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1622 -2.3046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7827 -2.1383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2369 -2.5925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5122 -3.0545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0310 -1.3465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5681 -0.8836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7376 -0.2513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3700 -0.0818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8328 -0.5447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6634 -1.1771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6637 -4.1658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1111 0.8841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6184 -1.9906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0837 -0.9410 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.3791 -1.4526 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.1890 -1.2902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7604 -1.4526 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.0559 -0.9410 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.4878 -1.1033 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.2778 -0.4074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5285 -0.6372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6237 1.7063 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-2.2848 1.2223 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.1919 1.8057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8032 2.2550 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.1421 2.7391 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.2351 2.1556 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.1894 1.7557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6402 2.3895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8479 3.0895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3686 -0.6786 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0487 -4.4726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0146 -4.7314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2308 -1.4371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9452 -1.8496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3160 -0.1218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0686 0.5367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6185 2.6879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9427 3.1610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
20 22 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
34 36 1 0 0 0 0
33 37 1 0 0 0 0
30 19 1 0 0 0 0
25 38 1 0 0 0 0
1 39 1 0 0 0 0
39 40 1 0 0 0 0
24 41 1 0 0 0 0
41 42 1 0 0 0 0
16 43 1 0 0 0 0
43 44 1 0 0 0 0
32 45 1 0 0 0 0
45 46 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 45 46
M SBL 4 1 49
M SMT 4 CH2OH
M SVB 4 49 3.2727 1.141
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 41 42
M SBL 3 1 45
M SMT 3 CH2OH
M SVB 3 45 1.2308 -1.4371
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 47
M SMT 2 OCH3
M SVB 2 47 0.1462 1.7287
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 43
M SMT 1 OCH3
M SVB 1 43 -3.9871 0.3444
S SKP 8
ID FL5FCDGA0002
KNApSAcK_ID C00005610
NAME Rhamnazin 3-galactoside-4'-glucoside
CAS_RN 106009-50-1
FORMULA C29H34O17
EXACTMASS 654.179599662
AVERAGEMASS 654.57006
SMILES C(C5O)([C@@H](O[C@@H]([C@@H]5O)CO)Oc(c4OC)ccc(c4)C(=C2O[C@H](O3)C(O)C([C@@H](O)[C@H]3CO)O)Oc(c1C2=O)cc(cc1O)OC)O
M END