Mol:FL5FEAGS0010
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3388 0.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3388 0.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3388 -0.3539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3388 -0.3539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6378 -0.7587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6378 -0.7587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0632 -0.3539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0632 -0.3539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0632 0.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0632 0.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6378 0.8602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6378 0.8602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7643 -0.7587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7643 -0.7587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4653 -0.3539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4653 -0.3539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4653 0.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4653 0.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7643 0.8602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7643 0.8602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7643 -1.6003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7643 -1.6003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1660 0.8601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1660 0.8601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8806 0.4475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8806 0.4475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5951 0.8601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5951 0.8601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5951 1.6851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5951 1.6851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8806 2.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8806 2.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1660 1.6851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1660 1.6851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0396 0.8601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0396 0.8601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2474 -0.8360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2474 -0.8360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2219 2.0471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2219 2.0471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6378 -1.5678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6378 -1.5678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0356 -2.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0356 -2.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8272 -2.7495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8272 -2.7495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5759 -1.8706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5759 -1.8706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8272 -0.9863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8272 -0.9863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0356 -0.5292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0356 -0.5292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2866 -1.4081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2866 -1.4081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2574 -1.4081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2574 -1.4081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1165 -2.9908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1165 -2.9908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6513 -3.4362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6513 -3.4362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1734 -2.4057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1734 -2.4057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5900 1.1692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5900 1.1692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7984 0.7121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7984 0.7121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0496 1.5911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0496 1.5911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7984 2.4753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7984 2.4753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5900 2.9325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5900 2.9325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3389 2.0535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3389 2.0535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2898 3.4362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2898 3.4362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7984 3.0458 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7984 3.0458 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1115 2.6967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1115 2.6967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2574 1.1742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2574 1.1742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9884 -0.7289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9884 -0.7289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0880 -1.2487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0880 -1.2487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 26 19 1 0 0 0 0 | + | 26 19 1 0 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 32 1 1 0 0 0 | + | 37 32 1 1 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 32 41 1 0 0 0 0 | + | 32 41 1 0 0 0 0 |
| − | 33 18 1 0 0 0 0 | + | 33 18 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 2 42 1 0 0 0 0 | + | 2 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 ^OCH3 | + | M SMT 1 ^OCH3 |
| − | M SBV 1 47 0.6496 0.3750 | + | M SBV 1 47 0.6496 0.3750 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FEAGS0010 | + | ID FL5FEAGS0010 |
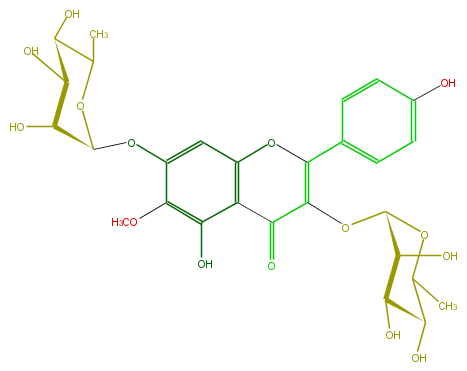
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES O=C(C=1OC(C5O)OC(C)C(C5O)O)c(c3O)c(cc(OC(C4O)OC(C(O)C4O)C)c3OC)OC1c(c2)ccc(O)c2 | + | SMILES O=C(C=1OC(C5O)OC(C)C(C5O)O)c(c3O)c(cc(OC(C4O)OC(C(O)C4O)C)c3OC)OC1c(c2)ccc(O)c2 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-1.3388 0.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3388 -0.3539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6378 -0.7587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0632 -0.3539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0632 0.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6378 0.8602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7643 -0.7587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4653 -0.3539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4653 0.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7643 0.8602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7643 -1.6003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1660 0.8601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8806 0.4475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5951 0.8601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5951 1.6851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8806 2.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1660 1.6851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0396 0.8601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2474 -0.8360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2219 2.0471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6378 -1.5678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0356 -2.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8272 -2.7495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5759 -1.8706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8272 -0.9863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0356 -0.5292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2866 -1.4081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2574 -1.4081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1165 -2.9908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6513 -3.4362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1734 -2.4057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5900 1.1692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7984 0.7121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0496 1.5911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7984 2.4753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5900 2.9325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3389 2.0535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2898 3.4362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7984 3.0458 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1115 2.6967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2574 1.1742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9884 -0.7289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0880 -1.2487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
20 15 1 0 0 0 0
3 21 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
27 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
26 19 1 0 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 1 0 0 0
37 32 1 1 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
37 40 1 0 0 0 0
32 41 1 0 0 0 0
33 18 1 0 0 0 0
42 43 1 0 0 0 0
2 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 47
M SMT 1 ^OCH3
M SBV 1 47 0.6496 0.3750
S SKP 5
ID FL5FEAGS0010
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES O=C(C=1OC(C5O)OC(C)C(C5O)O)c(c3O)c(cc(OC(C4O)OC(C(O)C4O)C)c3OC)OC1c(c2)ccc(O)c2
M END