Mol:FL5FECGS0034
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 38 41 0 0 0 0 0 0 0 0999 V2000 | + | 38 41 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.4104 -4.0952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4104 -4.0952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0818 -4.5081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0818 -4.5081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6854 -4.2884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6854 -4.2884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7969 -3.6558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7969 -3.6558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3048 -3.2429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3048 -3.2429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2988 -3.4626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2988 -3.4626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4006 -3.4361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4006 -3.4361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5121 -2.8035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5121 -2.8035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0200 -2.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0200 -2.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4163 -2.6103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4163 -2.6103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7842 -3.7580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7842 -3.7580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1315 -1.7582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1315 -1.7582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7467 -1.5343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7467 -1.5343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8604 -0.8895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8604 -0.8895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3589 -0.4687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3589 -0.4687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7437 -0.6928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7437 -0.6928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6300 -1.3374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6300 -1.3374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1773 -4.7012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1773 -4.7012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4305 1.2034 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.4305 1.2034 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.1009 0.5339 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.1009 0.5339 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7082 0.8477 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7082 0.8477 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2901 0.6823 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.2901 0.6823 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.7096 1.2557 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.7096 1.2557 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.0125 1.0381 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.0125 1.0381 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.8718 1.0537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8718 1.0537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2507 1.4507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2507 1.4507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4771 2.1237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4771 2.1237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0017 0.2973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0017 0.2973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4845 -0.1054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4845 -0.1054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0809 0.8095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0809 0.8095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4518 -2.4615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4518 -2.4615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3915 -2.1195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3915 -2.1195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1139 -3.9711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1139 -3.9711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0987 -3.7976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0987 -3.7976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1207 -5.3026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1207 -5.3026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6546 -5.5848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6546 -5.5848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5946 0.5018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5946 0.5018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9433 -0.2459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9433 -0.2459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 28 1 0 0 0 0 | + | 20 28 1 0 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 16 29 1 0 0 0 0 | + | 16 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 8 31 1 0 0 0 0 | + | 8 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 1 33 1 0 0 0 0 | + | 1 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 2 35 1 0 0 0 0 | + | 2 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 22 37 1 0 0 0 0 | + | 22 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 37 38 | + | M SAL 5 2 37 38 |
| − | M SBL 5 1 40 | + | M SBL 5 1 40 |
| − | M SMT 5 CH2OH | + | M SMT 5 CH2OH |
| − | M SVB 5 40 3.6431 0.7091 | + | M SVB 5 40 3.6431 0.7091 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 35 36 | + | M SAL 4 2 35 36 |
| − | M SBL 4 1 38 | + | M SBL 4 1 38 |
| − | M SMT 4 OCH3 | + | M SMT 4 OCH3 |
| − | M SVB 4 38 -3.6431 -1.7009 | + | M SVB 4 38 -3.6431 -1.7009 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 33 34 | + | M SAL 3 2 33 34 |
| − | M SBL 3 1 36 | + | M SBL 3 1 36 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 36 -3.4167 0.1007 | + | M SVB 3 36 -3.4167 0.1007 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 31 32 | + | M SAL 2 2 31 32 |
| − | M SBL 2 1 34 | + | M SBL 2 1 34 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 34 -0.6354 -1.3666 | + | M SVB 2 34 -0.6354 -1.3666 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 29 30 | + | M SAL 1 2 29 30 |
| − | M SBL 1 1 32 | + | M SBL 1 1 32 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 32 0.5722 1.3612 | + | M SVB 1 32 0.5722 1.3612 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FECGS0034 | + | ID FL5FECGS0034 |
| − | KNApSAcK_ID C00005681 | + | KNApSAcK_ID C00005681 |
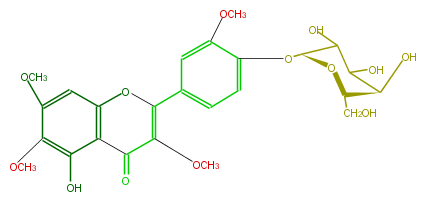
| − | NAME Galactobuxin | + | NAME Galactobuxin |
| − | CAS_RN 133362-61-5 | + | CAS_RN 133362-61-5 |
| − | FORMULA C25H28O13 | + | FORMULA C25H28O13 |
| − | EXACTMASS 536.152990982 | + | EXACTMASS 536.152990982 |
| − | AVERAGEMASS 536.48202 | + | AVERAGEMASS 536.48202 |
| − | SMILES C(C(OC)=2)(=O)c(c(OC2c(c3)ccc(O[C@@H](C4O)O[C@@H]([C@@H](C(O)4)O)CO)c(OC)3)1)c(O)c(OC)c(c1)OC | + | SMILES C(C(OC)=2)(=O)c(c(OC2c(c3)ccc(O[C@@H](C4O)O[C@@H]([C@@H](C(O)4)O)CO)c(OC)3)1)c(O)c(OC)c(c1)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
38 41 0 0 0 0 0 0 0 0999 V2000
-0.4104 -4.0952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0818 -4.5081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6854 -4.2884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7969 -3.6558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3048 -3.2429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2988 -3.4626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4006 -3.4361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5121 -2.8035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0200 -2.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4163 -2.6103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7842 -3.7580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1315 -1.7582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7467 -1.5343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8604 -0.8895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3589 -0.4687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7437 -0.6928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6300 -1.3374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1773 -4.7012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4305 1.2034 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.1009 0.5339 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7082 0.8477 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2901 0.6823 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.7096 1.2557 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.0125 1.0381 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.8718 1.0537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2507 1.4507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4771 2.1237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0017 0.2973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4845 -0.1054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0809 0.8095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4518 -2.4615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3915 -2.1195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1139 -3.9711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0987 -3.7976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1207 -5.3026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6546 -5.5848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5946 0.5018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9433 -0.2459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 28 1 0 0 0 0
15 28 1 0 0 0 0
16 29 1 0 0 0 0
29 30 1 0 0 0 0
8 31 1 0 0 0 0
31 32 1 0 0 0 0
1 33 1 0 0 0 0
33 34 1 0 0 0 0
2 35 1 0 0 0 0
35 36 1 0 0 0 0
22 37 1 0 0 0 0
37 38 1 0 0 0 0
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 37 38
M SBL 5 1 40
M SMT 5 CH2OH
M SVB 5 40 3.6431 0.7091
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 35 36
M SBL 4 1 38
M SMT 4 OCH3
M SVB 4 38 -3.6431 -1.7009
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 33 34
M SBL 3 1 36
M SMT 3 OCH3
M SVB 3 36 -3.4167 0.1007
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 31 32
M SBL 2 1 34
M SMT 2 OCH3
M SVB 2 34 -0.6354 -1.3666
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 29 30
M SBL 1 1 32
M SMT 1 OCH3
M SVB 1 32 0.5722 1.3612
S SKP 8
ID FL5FECGS0034
KNApSAcK_ID C00005681
NAME Galactobuxin
CAS_RN 133362-61-5
FORMULA C25H28O13
EXACTMASS 536.152990982
AVERAGEMASS 536.48202
SMILES C(C(OC)=2)(=O)c(c(OC2c(c3)ccc(O[C@@H](C4O)O[C@@H]([C@@H](C(O)4)O)CO)c(OC)3)1)c(O)c(OC)c(c1)OC
M END