Mol:FL63ACGS0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 61 0 0 0 0 0 0 0 0999 V2000 | + | 56 61 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.0672 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0672 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0672 1.0409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0672 1.0409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5852 0.7419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5852 0.7419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1032 1.0409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1032 1.0409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1032 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1032 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5852 1.9381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5852 1.9381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6212 0.7419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6212 0.7419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1392 1.0409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1392 1.0409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1392 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1392 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6212 1.9381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6212 1.9381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6572 1.9381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6572 1.9381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1795 1.6366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1795 1.6366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7018 1.9381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7018 1.9381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7018 2.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7018 2.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1795 2.8428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1795 2.8428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6572 2.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6572 2.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4491 2.9727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4491 2.9727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4115 -0.1377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4115 -0.1377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0403 -0.6276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0403 -0.6276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5058 -0.4198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5058 -0.4198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0100 -0.4142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0100 -0.4142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3648 -0.0394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3648 -0.0394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8324 -0.2861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8324 -0.2861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9253 -0.4343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9253 -0.4343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6189 -0.6558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6189 -0.6558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1995 -0.9339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1995 -0.9339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3349 0.2163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3349 0.2163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5852 0.1451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5852 0.1451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4507 1.9381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4507 1.9381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6572 0.7419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6572 0.7419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1795 3.4450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1795 3.4450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1713 0.8270 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1713 0.8270 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4259 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4259 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1515 1.7433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1515 1.7433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9745 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9745 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2488 0.7929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2488 0.7929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7975 0.7929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7975 0.7929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0729 1.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0729 1.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6237 1.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6237 1.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8991 0.7929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8991 0.7929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6237 0.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6237 0.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0729 0.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0729 0.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4491 0.7929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4491 0.7929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0363 -1.5432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0363 -1.5432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3060 -2.0104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3060 -2.0104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6037 -1.5432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6037 -1.5432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9234 -2.0969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9234 -2.0969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5627 -2.0969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5627 -2.0969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8648 -2.6202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8648 -2.6202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4690 -2.6202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4690 -2.6202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7711 -2.0969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7711 -2.0969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4690 -1.5737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4690 -1.5737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8648 -1.5737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8648 -1.5737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3747 -2.0969 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3747 -2.0969 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4687 -3.0325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4687 -3.0325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1832 -3.4450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1832 -3.4450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 6 0 0 0 | + | 9 11 1 6 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 17 14 1 0 0 0 0 | + | 17 14 1 0 0 0 0 |
| − | 18 19 1 1 0 0 0 | + | 18 19 1 1 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 21 20 1 1 0 0 0 | + | 21 20 1 1 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 18 1 0 0 0 0 | + | 23 18 1 0 0 0 0 |
| − | 18 24 1 0 0 0 0 | + | 18 24 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 3 28 1 0 0 0 0 | + | 3 28 1 0 0 0 0 |
| − | 28 21 1 0 0 0 0 | + | 28 21 1 0 0 0 0 |
| − | 1 29 1 0 0 0 0 | + | 1 29 1 0 0 0 0 |
| − | 8 30 1 1 0 0 0 | + | 8 30 1 1 0 0 0 |
| − | 15 31 1 0 0 0 0 | + | 15 31 1 0 0 0 0 |
| − | 27 32 1 0 0 0 0 | + | 27 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 42 37 1 0 0 0 0 | + | 42 37 1 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | 26 44 1 0 0 0 0 | + | 26 44 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 46 47 2 0 0 0 0 | + | 46 47 2 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 2 0 0 0 0 | + | 48 49 2 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 52 53 2 0 0 0 0 | + | 52 53 2 0 0 0 0 |
| − | 53 48 1 0 0 0 0 | + | 53 48 1 0 0 0 0 |
| − | 51 54 1 0 0 0 0 | + | 51 54 1 0 0 0 0 |
| − | 50 55 1 0 0 0 0 | + | 50 55 1 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 55 56 | + | M SAL 1 2 55 56 |
| − | M SBL 1 1 60 | + | M SBL 1 1 60 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 60 -5.8895 7.6204 | + | M SBV 1 60 -5.8895 7.6204 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL63ACGS0012 | + | ID FL63ACGS0012 |
| − | KNApSAcK_ID C00008848 | + | KNApSAcK_ID C00008848 |
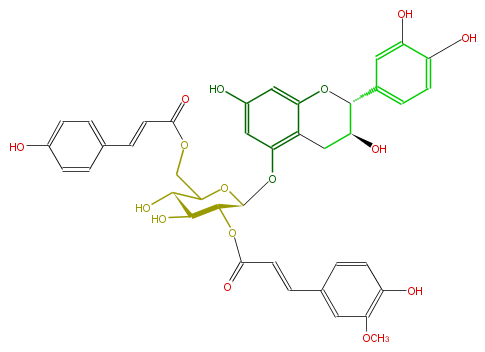
| − | NAME Catechin 5-O-(2-feruloyl-6-p-coumaroyl-beta-D-glucopyranoside) | + | NAME Catechin 5-O-(2-feruloyl-6-p-coumaroyl-beta-D-glucopyranoside) |
| − | CAS_RN 103215-60-7 | + | CAS_RN 103215-60-7 |
| − | FORMULA C40H38O16 | + | FORMULA C40H38O16 |
| − | EXACTMASS 774.2159851680001 | + | EXACTMASS 774.2159851680001 |
| − | AVERAGEMASS 774.7201200000001 | + | AVERAGEMASS 774.7201200000001 |
| − | SMILES c(O)(c1)c(OC)cc(C=CC(OC(C3Oc(c64)cc(O)cc4OC(C(C6)O)c(c5)cc(O)c(O)c5)C(C(O)C(O3)COC(C=Cc(c2)ccc(O)c2)=O)O)=O)c1 | + | SMILES c(O)(c1)c(OC)cc(C=CC(OC(C3Oc(c64)cc(O)cc4OC(C(C6)O)c(c5)cc(O)c(O)c5)C(C(O)C(O3)COC(C=Cc(c2)ccc(O)c2)=O)O)=O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 61 0 0 0 0 0 0 0 0999 V2000
0.0672 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0672 1.0409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5852 0.7419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1032 1.0409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1032 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5852 1.9381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6212 0.7419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1392 1.0409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1392 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6212 1.9381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6572 1.9381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1795 1.6366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7018 1.9381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7018 2.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1795 2.8428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6572 2.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4491 2.9727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4115 -0.1377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0403 -0.6276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5058 -0.4198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0100 -0.4142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3648 -0.0394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8324 -0.2861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9253 -0.4343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6189 -0.6558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1995 -0.9339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3349 0.2163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5852 0.1451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4507 1.9381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6572 0.7419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1795 3.4450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1713 0.8270 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4259 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1515 1.7433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9745 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2488 0.7929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7975 0.7929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0729 1.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6237 1.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8991 0.7929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6237 0.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0729 0.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4491 0.7929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0363 -1.5432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3060 -2.0104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6037 -1.5432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9234 -2.0969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5627 -2.0969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8648 -2.6202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4690 -2.6202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7711 -2.0969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4690 -1.5737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8648 -1.5737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3747 -2.0969 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4687 -3.0325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1832 -3.4450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 6 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
17 14 1 0 0 0 0
18 19 1 1 0 0 0
19 20 1 1 0 0 0
21 20 1 1 0 0 0
21 22 1 0 0 0 0
22 23 1 0 0 0 0
23 18 1 0 0 0 0
18 24 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
23 27 1 0 0 0 0
3 28 1 0 0 0 0
28 21 1 0 0 0 0
1 29 1 0 0 0 0
8 30 1 1 0 0 0
15 31 1 0 0 0 0
27 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
42 37 1 0 0 0 0
40 43 1 0 0 0 0
26 44 1 0 0 0 0
44 45 2 0 0 0 0
44 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 52 1 0 0 0 0
52 53 2 0 0 0 0
53 48 1 0 0 0 0
51 54 1 0 0 0 0
50 55 1 0 0 0 0
55 56 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 55 56
M SBL 1 1 60
M SMT 1 OCH3
M SBV 1 60 -5.8895 7.6204
S SKP 8
ID FL63ACGS0012
KNApSAcK_ID C00008848
NAME Catechin 5-O-(2-feruloyl-6-p-coumaroyl-beta-D-glucopyranoside)
CAS_RN 103215-60-7
FORMULA C40H38O16
EXACTMASS 774.2159851680001
AVERAGEMASS 774.7201200000001
SMILES c(O)(c1)c(OC)cc(C=CC(OC(C3Oc(c64)cc(O)cc4OC(C(C6)O)c(c5)cc(O)c(O)c5)C(C(O)C(O3)COC(C=Cc(c2)ccc(O)c2)=O)O)=O)c1
M END