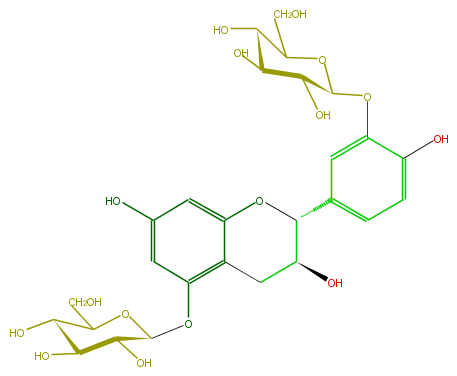
Mol:FL63ACGS0021
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3400 -0.6562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3400 -0.6562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3400 -1.4811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3400 -1.4811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6255 -1.8936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6255 -1.8936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0889 -1.4811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0889 -1.4811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0889 -0.6562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0889 -0.6562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6255 -0.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6255 -0.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8034 -1.8936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8034 -1.8936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5178 -1.4811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5178 -1.4811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5178 -0.6562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5178 -0.6562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8034 -0.2436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8034 -0.2436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2323 -0.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2323 -0.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9527 -0.6596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9527 -0.6596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6730 -0.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6730 -0.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6730 0.5882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6730 0.5882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9527 1.0040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9527 1.0040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2323 0.5882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2323 0.5882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3625 0.9862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3625 0.9862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0544 -0.2436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0544 -0.2436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2323 -1.8936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2323 -1.8936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9527 1.8348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9527 1.8348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6255 -2.7166 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6255 -2.7166 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7368 3.1831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7368 3.1831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8421 2.3419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8421 2.3419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6239 2.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6239 2.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2439 1.8725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2439 1.8725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0546 2.5787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0546 2.5787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3260 2.6064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3260 2.6064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0958 3.1782 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0958 3.1782 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3598 2.7059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3598 2.7059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9884 1.4664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9884 1.4664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3350 -2.6363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3350 -2.6363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8230 -3.3120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8230 -3.3120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0858 -3.0254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0858 -3.0254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3745 -3.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3745 -3.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8913 -2.5006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8913 -2.5006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5363 -2.8410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5363 -2.8410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9759 -2.8815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9759 -2.8815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4753 -3.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4753 -3.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5781 -3.4897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5781 -3.4897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0462 -2.2748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0462 -2.2748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3625 -1.8631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3625 -1.8631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0538 3.4897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0538 3.4897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0392 2.9207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0392 2.9207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 6 0 0 0 | + | 9 11 1 6 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 17 14 1 0 0 0 0 | + | 17 14 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 1 0 0 0 | + | 8 19 1 1 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 21 1 0 0 0 0 | + | 34 21 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 27 42 1 0 0 0 0 | + | 27 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 45 0.5099 -0.5661 | + | M SBV 1 45 0.5099 -0.5661 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 47 0.2722 -0.8833 | + | M SBV 2 47 0.2722 -0.8833 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL63ACGS0021 | + | ID FL63ACGS0021 |
| − | FORMULA C27H34O16 | + | FORMULA C27H34O16 |
| − | EXACTMASS 614.18468504 | + | EXACTMASS 614.18468504 |
| − | AVERAGEMASS 614.54926 | + | AVERAGEMASS 614.54926 |
| − | SMILES c(c14)c(cc(OC(C5O)OC(CO)C(O)C5O)c(CC(C(O4)c(c2)ccc(O)c2OC(O3)C(C(C(O)C(CO)3)O)O)O)1)O | + | SMILES c(c14)c(cc(OC(C5O)OC(CO)C(O)C5O)c(CC(C(O4)c(c2)ccc(O)c2OC(O3)C(C(C(O)C(CO)3)O)O)O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-1.3400 -0.6562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3400 -1.4811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6255 -1.8936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0889 -1.4811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0889 -0.6562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6255 -0.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8034 -1.8936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5178 -1.4811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5178 -0.6562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8034 -0.2436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2323 -0.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9527 -0.6596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6730 -0.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6730 0.5882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9527 1.0040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2323 0.5882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3625 0.9862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0544 -0.2436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2323 -1.8936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9527 1.8348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6255 -2.7166 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7368 3.1831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8421 2.3419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6239 2.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2439 1.8725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0546 2.5787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3260 2.6064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0958 3.1782 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3598 2.7059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9884 1.4664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3350 -2.6363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8230 -3.3120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0858 -3.0254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3745 -3.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8913 -2.5006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5363 -2.8410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9759 -2.8815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4753 -3.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5781 -3.4897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0462 -2.2748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3625 -1.8631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0538 3.4897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0392 2.9207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 6 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
17 14 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 1 0 0 0
15 20 1 0 0 0 0
3 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 20 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 21 1 0 0 0 0
40 41 1 0 0 0 0
36 40 1 0 0 0 0
42 43 1 0 0 0 0
27 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^ CH2OH
M SBV 1 45 0.5099 -0.5661
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 ^ CH2OH
M SBV 2 47 0.2722 -0.8833
S SKP 5
ID FL63ACGS0021
FORMULA C27H34O16
EXACTMASS 614.18468504
AVERAGEMASS 614.54926
SMILES c(c14)c(cc(OC(C5O)OC(CO)C(O)C5O)c(CC(C(O4)c(c2)ccc(O)c2OC(O3)C(C(C(O)C(CO)3)O)O)O)1)O
M END